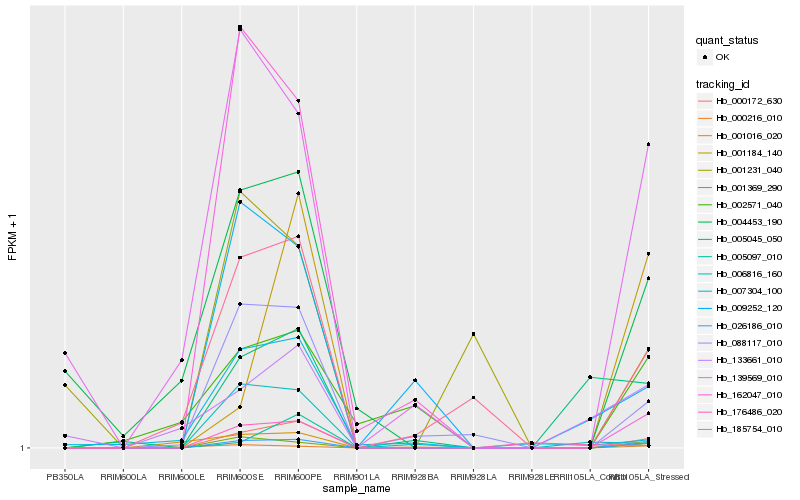
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026186_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06344 [Jatropha curcas] |
| 2 |
Hb_000172_630 |
0.1039875792 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 3 |
Hb_001369_290 |
0.1558481213 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_088117_010 |
0.2249237657 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_139569_010 |
0.2391066079 |
- |
- |
hypothetical protein JCGZ_24635 [Jatropha curcas] |
| 6 |
Hb_185754_010 |
0.2497071854 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_004453_190 |
0.2765327475 |
- |
- |
- |
| 8 |
Hb_001016_020 |
0.2771630541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009252_120 |
0.3133207353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005097_010 |
0.3231637295 |
- |
- |
PREDICTED: aluminum-activated malate transporter 7-like isoform X1 [Populus euphratica] |
| 11 |
Hb_006816_160 |
0.3235180607 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 12 |
Hb_133661_010 |
0.3334574376 |
- |
- |
hypothetical protein JCGZ_11734 [Jatropha curcas] |
| 13 |
Hb_001184_140 |
0.3356349225 |
- |
- |
hypothetical protein B456_008G022200 [Gossypium raimondii] |
| 14 |
Hb_002571_040 |
0.3392901883 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 15 |
Hb_007304_100 |
0.3471061527 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 16 |
Hb_005045_050 |
0.3471598705 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 17 |
Hb_162047_010 |
0.347831588 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 18 |
Hb_001231_040 |
0.3500334374 |
- |
- |
hypothetical protein POPTR_0006s06470g [Populus trichocarpa] |
| 19 |
Hb_176486_020 |
0.352956247 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Populus euphratica] |
| 20 |
Hb_000216_010 |
0.3551519524 |
- |
- |
carbon monoxide dehydrogenase [Streptomyces sp. CNQ-509] |