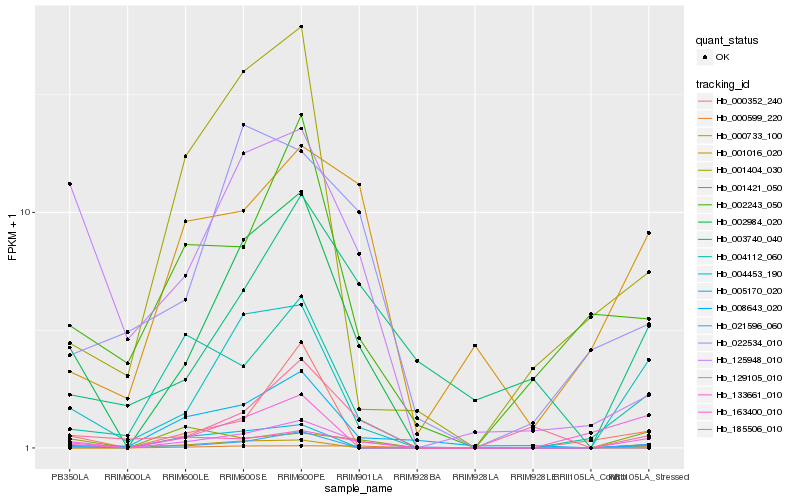
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129105_010 |
0.0 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_004453_190 |
0.1798152222 |
- |
- |
- |
| 3 |
Hb_004112_060 |
0.2562848096 |
- |
- |
hypothetical protein POPTR_0017s01950g, partial [Populus trichocarpa] |
| 4 |
Hb_002984_020 |
0.2775348845 |
- |
- |
hypothetical protein EUGRSUZ_J02661 [Eucalyptus grandis] |
| 5 |
Hb_000733_100 |
0.2801378869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000599_220 |
0.2837527022 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 7 |
Hb_185506_010 |
0.2940643554 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_002243_050 |
0.2949069976 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001421_050 |
0.2999935417 |
- |
- |
- |
| 10 |
Hb_125948_010 |
0.3036877121 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_021596_060 |
0.3040885877 |
- |
- |
PREDICTED: peroxidase 56-like, partial [Solanum tuberosum] |
| 12 |
Hb_005170_020 |
0.3059165703 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 13 |
Hb_003740_040 |
0.3066041575 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 14 |
Hb_001016_020 |
0.3086315941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_163400_010 |
0.3092949811 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 16 |
Hb_000352_240 |
0.3148087131 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_022534_010 |
0.3166355596 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 18 |
Hb_008643_020 |
0.3171749673 |
- |
- |
Cf-4/9 disease resistance-like family protein [Populus trichocarpa] |
| 19 |
Hb_001404_030 |
0.3188192826 |
- |
- |
DWNN domain isoform 3 [Theobroma cacao] |
| 20 |
Hb_133661_010 |
0.3195258166 |
- |
- |
hypothetical protein JCGZ_11734 [Jatropha curcas] |