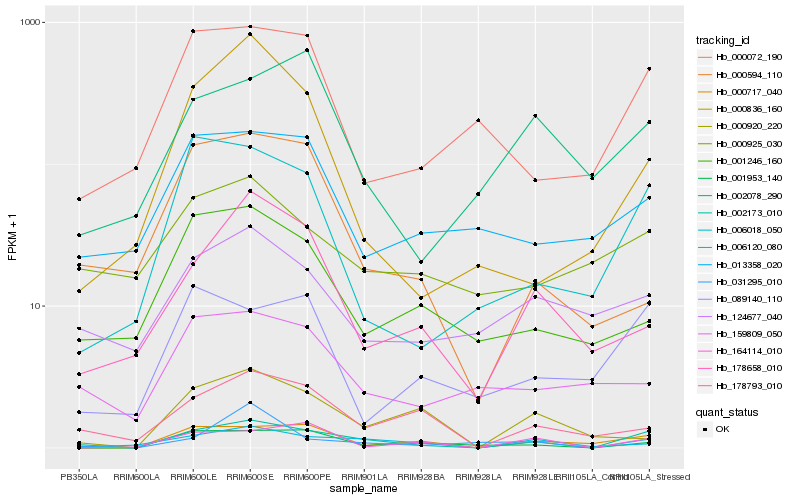
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002173_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_006120_080 |
0.2564646357 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 3 |
Hb_000717_040 |
0.2585440229 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0015s03590g [Populus trichocarpa] |
| 4 |
Hb_006018_050 |
0.2645362948 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 5 |
Hb_178793_010 |
0.2682175225 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Nelumbo nucifera] |
| 6 |
Hb_001953_140 |
0.2714101209 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000072_190 |
0.2820956436 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 8 |
Hb_164114_010 |
0.2874909014 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_178658_010 |
0.2878834958 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 10 |
Hb_089140_110 |
0.2948985578 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_124677_040 |
0.2962455542 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 12 |
Hb_002078_290 |
0.2990372502 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 13 |
Hb_013358_020 |
0.2990563537 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 14 |
Hb_001246_160 |
0.300530657 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 15 |
Hb_000836_160 |
0.3020479207 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 16 |
Hb_031295_010 |
0.3029942828 |
- |
- |
hypothetical protein JCGZ_19530 [Jatropha curcas] |
| 17 |
Hb_159809_050 |
0.30368187 |
- |
- |
AT14A, putative [Ricinus communis] |
| 18 |
Hb_000920_220 |
0.3041588813 |
- |
- |
PREDICTED: uncharacterized protein LOC105631216 [Jatropha curcas] |
| 19 |
Hb_000925_030 |
0.3044526105 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000594_110 |
0.3049048716 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |