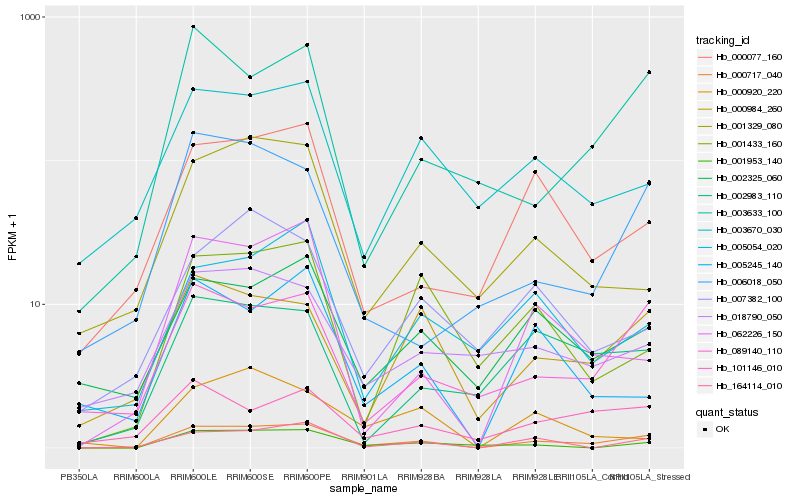
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000717_040 |
0.0 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0015s03590g [Populus trichocarpa] |
| 2 |
Hb_164114_010 |
0.1909533261 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_089140_110 |
0.2013464769 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002325_060 |
0.2106439678 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 5 |
Hb_000984_260 |
0.2115705466 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 6 |
Hb_001953_140 |
0.2119762599 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 7 |
Hb_003633_100 |
0.216283766 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |
| 8 |
Hb_062226_150 |
0.2194708155 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000077_160 |
0.2209928245 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 10 |
Hb_002983_110 |
0.2230689892 |
- |
- |
PREDICTED: uncharacterized protein LOC105634780 [Jatropha curcas] |
| 11 |
Hb_005054_020 |
0.224763556 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 12 |
Hb_003670_030 |
0.2297554319 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 13 |
Hb_001433_160 |
0.2343251827 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 14 |
Hb_018790_050 |
0.2344046177 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 15 |
Hb_101146_010 |
0.2357160487 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 16 |
Hb_006018_050 |
0.2367797831 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 17 |
Hb_001329_080 |
0.238730661 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 18 |
Hb_005245_140 |
0.2420639342 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 19 |
Hb_007382_100 |
0.2423646919 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000920_220 |
0.2436927327 |
- |
- |
PREDICTED: uncharacterized protein LOC105631216 [Jatropha curcas] |