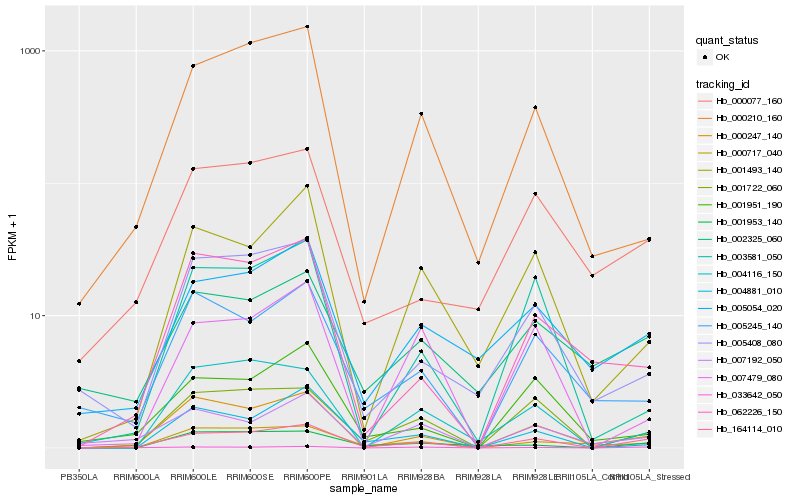
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164114_010 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001722_060 |
0.1574781053 |
- |
- |
- |
| 3 |
Hb_000717_040 |
0.1909533261 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0015s03590g [Populus trichocarpa] |
| 4 |
Hb_005054_020 |
0.2007455939 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 5 |
Hb_033642_050 |
0.2018631831 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 4-like [Jatropha curcas] |
| 6 |
Hb_000077_160 |
0.2079722292 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 7 |
Hb_001493_140 |
0.2120238536 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 8 |
Hb_007192_050 |
0.2121632972 |
- |
- |
PREDICTED: uncharacterized protein LOC105640295 [Jatropha curcas] |
| 9 |
Hb_001951_190 |
0.2156864283 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000247_140 |
0.2165496341 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 11 |
Hb_001953_140 |
0.2216331278 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000210_160 |
0.2222910923 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 13 |
Hb_003581_050 |
0.222989915 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 14 |
Hb_005408_080 |
0.2231476234 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 15 |
Hb_004116_150 |
0.2235530554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_062226_150 |
0.2241370617 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002325_060 |
0.2242292956 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 18 |
Hb_004881_010 |
0.2251198531 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 19 |
Hb_007479_080 |
0.2266852526 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005245_140 |
0.2269434182 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |