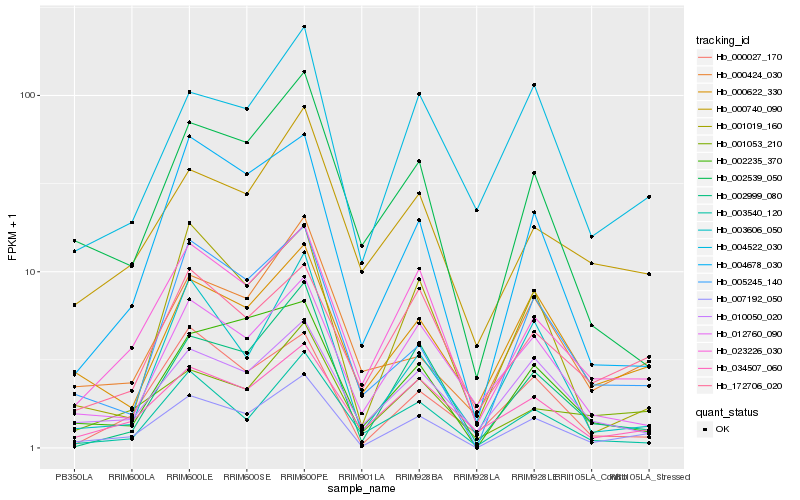
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007192_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640295 [Jatropha curcas] |
| 2 |
Hb_023226_030 |
0.1313827151 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 3 |
Hb_005245_140 |
0.1510518825 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |
| 4 |
Hb_004678_030 |
0.1529633717 |
- |
- |
fructokinase [Manihot esculenta] |
| 5 |
Hb_002999_080 |
0.1551189568 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 6 |
Hb_034507_060 |
0.1551365544 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 7 |
Hb_001053_210 |
0.1591206944 |
- |
- |
- |
| 8 |
Hb_000622_330 |
0.1592128494 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 9 |
Hb_002235_370 |
0.1640254615 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 10 |
Hb_010050_020 |
0.1642656394 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 11 |
Hb_004522_030 |
0.1652587286 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 12 |
Hb_000740_090 |
0.1659879676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003606_050 |
0.1687474908 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 14 |
Hb_001019_160 |
0.1688263944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003540_120 |
0.1691464014 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 16 |
Hb_000424_030 |
0.1693211427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_172706_020 |
0.1697369004 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 18 |
Hb_002539_050 |
0.1710279062 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 19 |
Hb_012760_090 |
0.1730974402 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 20 |
Hb_000027_170 |
0.173740312 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |