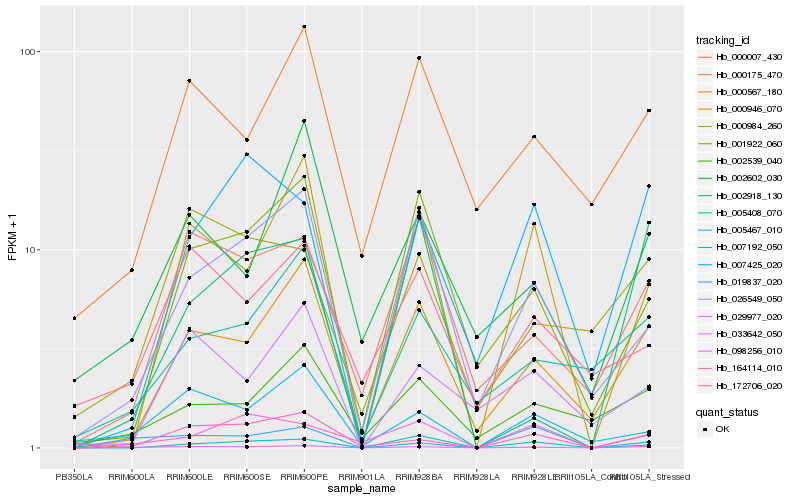
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033642_050 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 4-like [Jatropha curcas] |
| 2 |
Hb_164114_010 |
0.2018631831 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000567_180 |
0.2039220971 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 4 |
Hb_007192_050 |
0.2353774616 |
- |
- |
PREDICTED: uncharacterized protein LOC105640295 [Jatropha curcas] |
| 5 |
Hb_000946_070 |
0.2403039212 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 6 |
Hb_000007_430 |
0.2437839324 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 7 |
Hb_002539_040 |
0.2466256739 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_005408_070 |
0.2490026088 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 9 |
Hb_026549_050 |
0.2500914513 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000984_260 |
0.2585962609 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 11 |
Hb_001922_060 |
0.2592007829 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000175_470 |
0.2602852635 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_002602_030 |
0.2624217171 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 14 |
Hb_019837_020 |
0.2637719652 |
- |
- |
- |
| 15 |
Hb_007425_020 |
0.268087255 |
transcription factor |
TF Family: ERF |
PREDICTED: pathogenesis-related genes transcriptional activator PTI6-like [Jatropha curcas] |
| 16 |
Hb_002918_130 |
0.2690689481 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 17 |
Hb_029977_020 |
0.2721213183 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 18 |
Hb_172706_020 |
0.27229484 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 19 |
Hb_098256_010 |
0.2727029449 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 20 |
Hb_005467_010 |
0.273753055 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |