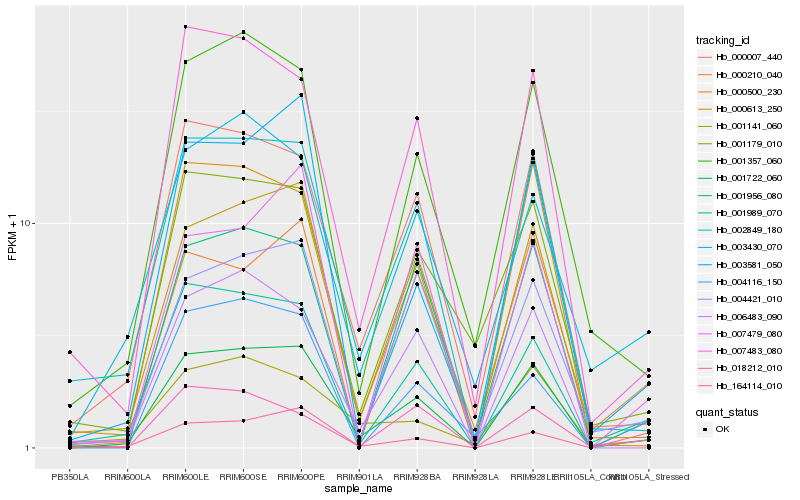
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001722_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_001989_070 |
0.153166087 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 3 |
Hb_002849_180 |
0.154950841 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 4 |
Hb_164114_010 |
0.1574781053 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000613_250 |
0.1589908292 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 6 |
Hb_000007_440 |
0.1603586886 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 7 |
Hb_003430_070 |
0.1624348006 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 8 |
Hb_000210_040 |
0.1661015247 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 9 |
Hb_007483_080 |
0.1672559657 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 10 |
Hb_001956_080 |
0.1675317381 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 11 |
Hb_001357_060 |
0.171014868 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 12 |
Hb_003581_050 |
0.172056379 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 13 |
Hb_004421_010 |
0.172856778 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 14 |
Hb_004116_150 |
0.1763108001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007479_080 |
0.1763278967 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 16 |
Hb_018212_010 |
0.1766186066 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 17 |
Hb_006483_090 |
0.1784216157 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 18 |
Hb_001179_010 |
0.1785263275 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 19 |
Hb_000500_230 |
0.1793781577 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 20 |
Hb_001141_060 |
0.1794139013 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |