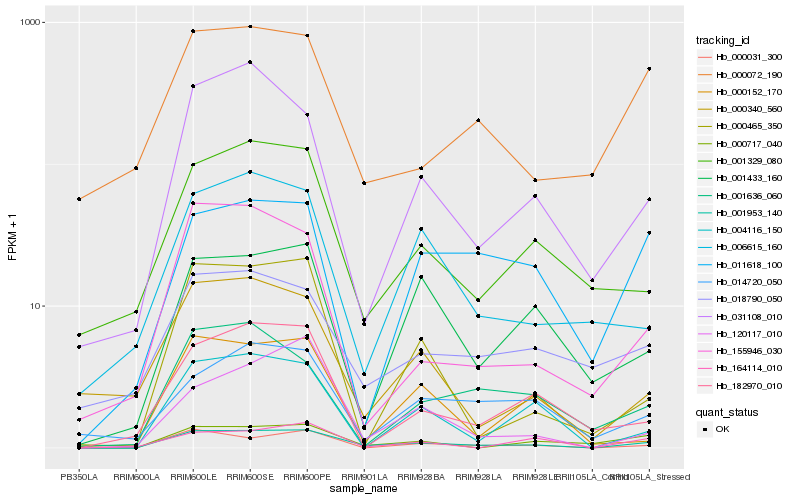
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001953_140 |
0.0 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000031_300 |
0.1806375883 |
- |
- |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 3 |
Hb_004116_150 |
0.199124118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_031108_010 |
0.200427348 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 5 |
Hb_001329_080 |
0.210304738 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 6 |
Hb_000717_040 |
0.2119762599 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0015s03590g [Populus trichocarpa] |
| 7 |
Hb_001433_160 |
0.2150199468 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 8 |
Hb_120117_010 |
0.2160059921 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_018790_050 |
0.2166153151 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 10 |
Hb_014720_050 |
0.2167637959 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 11 |
Hb_000152_170 |
0.2181067368 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 12 |
Hb_182970_010 |
0.220005325 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 13 |
Hb_155946_030 |
0.2209284444 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 14 |
Hb_000465_350 |
0.2211710869 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 2 [Jatropha curcas] |
| 15 |
Hb_011618_100 |
0.2213459619 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 16 |
Hb_164114_010 |
0.2216331278 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000072_190 |
0.2217117416 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 18 |
Hb_006615_160 |
0.2218463105 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 19 |
Hb_001636_060 |
0.2232672219 |
- |
- |
Putative serine/threonine-protein kinase-like protein CCR3 [Glycine soja] |
| 20 |
Hb_000340_560 |
0.2238328891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |