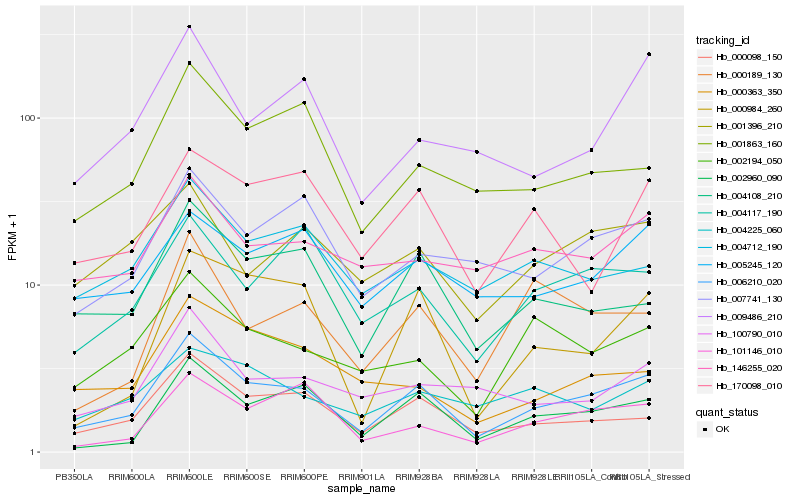
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006210_020 |
0.0 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000098_150 |
0.1309867072 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 3 |
Hb_002960_090 |
0.1416398183 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_004108_210 |
0.143202553 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 5 |
Hb_007741_130 |
0.1543656004 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 6 |
Hb_002194_050 |
0.1573095719 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004117_190 |
0.159528674 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 8 |
Hb_001863_160 |
0.1605741748 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 9 |
Hb_005245_120 |
0.1621098419 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_004712_190 |
0.1629907946 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 11 |
Hb_100790_010 |
0.1635825624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001396_210 |
0.1647369574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000984_260 |
0.1654980919 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 14 |
Hb_009486_210 |
0.1663571669 |
- |
- |
- |
| 15 |
Hb_101146_010 |
0.1664278474 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 16 |
Hb_170098_010 |
0.1674667146 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 17 |
Hb_146255_020 |
0.1681581144 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 18 |
Hb_000189_130 |
0.1692091483 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000363_350 |
0.1700780152 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_004225_060 |
0.1719656569 |
- |
- |
hypothetical protein CISIN_1g027511mg [Citrus sinensis] |