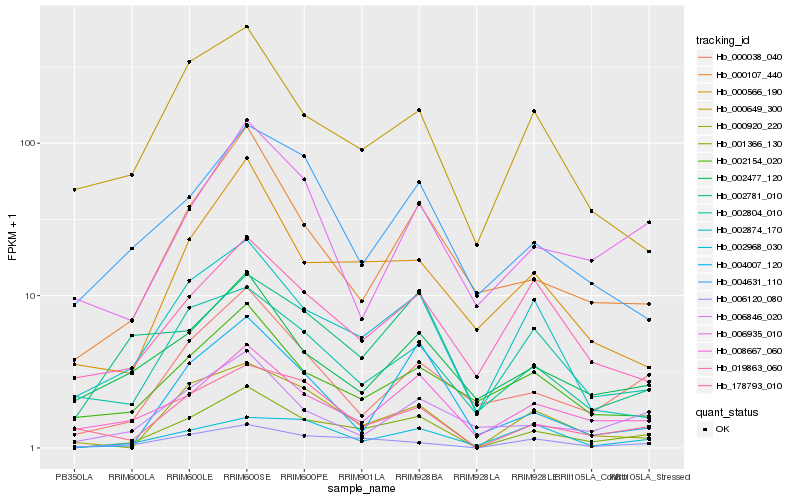
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_130 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_006120_080 |
0.1709334567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 3 |
Hb_000920_220 |
0.1865594703 |
- |
- |
PREDICTED: uncharacterized protein LOC105631216 [Jatropha curcas] |
| 4 |
Hb_008667_060 |
0.1907558048 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 5 |
Hb_178793_010 |
0.1933150611 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Nelumbo nucifera] |
| 6 |
Hb_002874_170 |
0.1945171074 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_000038_040 |
0.1971529251 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 8 |
Hb_000107_440 |
0.2007319364 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 9 |
Hb_000566_190 |
0.2032966572 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002477_120 |
0.2143388123 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_004007_120 |
0.2161699764 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type isoform X1 [Populus euphratica] |
| 12 |
Hb_000649_300 |
0.2167072983 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 13 |
Hb_006846_020 |
0.2175869651 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 14 |
Hb_002781_010 |
0.2175936085 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002154_020 |
0.2177249516 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_019863_060 |
0.2182763715 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 17 |
Hb_004631_110 |
0.2191996892 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_006935_010 |
0.2192248139 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |
| 19 |
Hb_002804_010 |
0.2197908444 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 20 |
Hb_002968_030 |
0.2198775025 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |