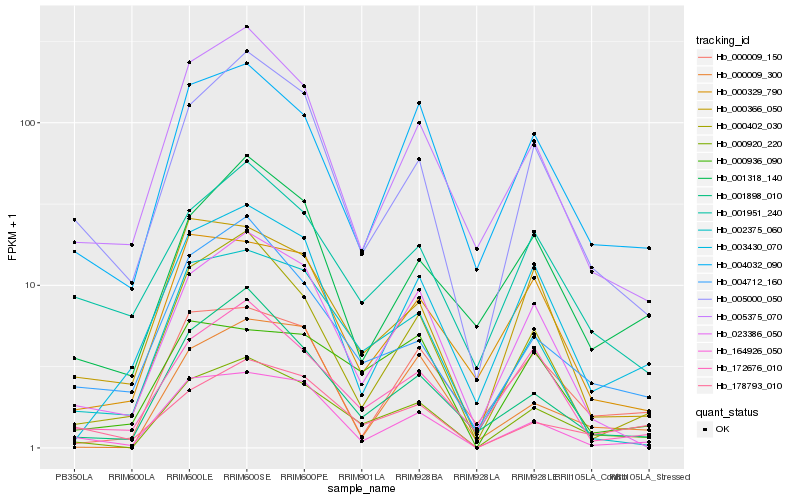
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631216 [Jatropha curcas] |
| 2 |
Hb_005000_050 |
0.152842015 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 3 |
Hb_164926_050 |
0.1537241544 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 4 |
Hb_004032_090 |
0.1579738452 |
- |
- |
PREDICTED: protein AIG1 [Jatropha curcas] |
| 5 |
Hb_023386_050 |
0.1596176975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001898_010 |
0.1613317648 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 7 |
Hb_002375_060 |
0.161504794 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 8 |
Hb_178793_010 |
0.1615404678 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Nelumbo nucifera] |
| 9 |
Hb_000402_030 |
0.1627585518 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 10 |
Hb_004712_160 |
0.1668501834 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 11 |
Hb_172676_010 |
0.1674723286 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 12 |
Hb_005375_070 |
0.1758323554 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 13 |
Hb_000009_150 |
0.1758884342 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 14 |
Hb_003430_070 |
0.1760419848 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 15 |
Hb_000366_050 |
0.1769161272 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_000009_300 |
0.1797846381 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |
| 17 |
Hb_000329_790 |
0.1825841631 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 18 |
Hb_000936_090 |
0.1836953805 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X2 [Jatropha curcas] |
| 19 |
Hb_001951_240 |
0.1839378106 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 20 |
Hb_001318_140 |
0.1842354639 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |