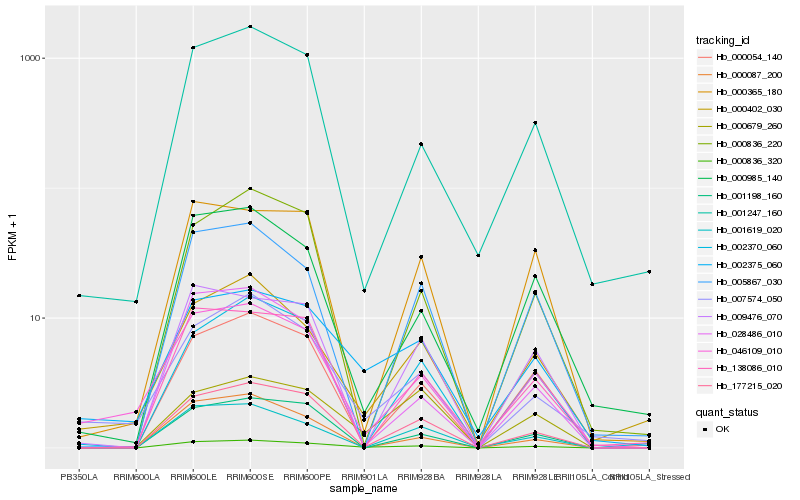
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_320 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_177215_020 |
0.1234002869 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 3 |
Hb_000054_140 |
0.1391437506 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 4 |
Hb_002375_060 |
0.1456111715 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 5 |
Hb_001198_160 |
0.1467606038 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_001619_020 |
0.1468717391 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 7 |
Hb_000985_140 |
0.1519070434 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 8 |
Hb_138086_010 |
0.1538170602 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 9 |
Hb_002370_060 |
0.1556880135 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 10 |
Hb_000836_220 |
0.1563723464 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000087_200 |
0.1575839966 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 12 |
Hb_001247_160 |
0.157887164 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 13 |
Hb_005867_030 |
0.1590543092 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 14 |
Hb_028486_010 |
0.1617535707 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 15 |
Hb_009476_070 |
0.1625845533 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 16 |
Hb_046109_010 |
0.1669815994 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 17 |
Hb_007574_050 |
0.1685155674 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 18 |
Hb_000365_180 |
0.1687511817 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 19 |
Hb_000402_030 |
0.1692142402 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_000679_260 |
0.1703374575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |