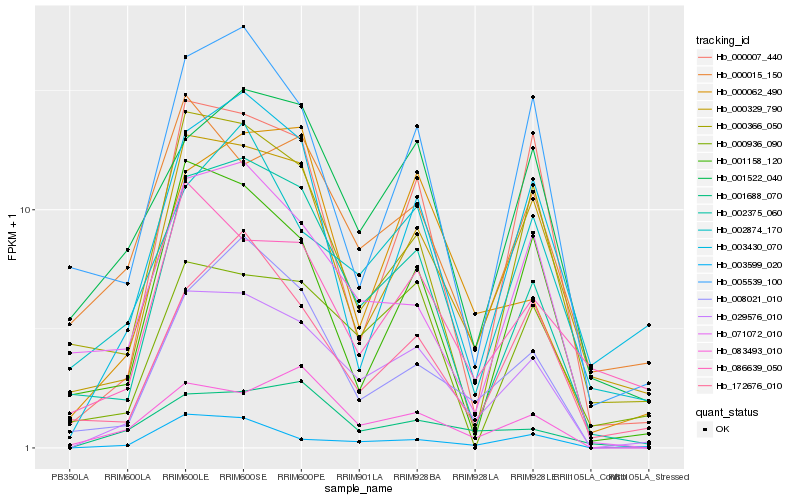
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029576_010 |
0.0 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31-like [Glycine max] |
| 2 |
Hb_002375_060 |
0.1397186753 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 3 |
Hb_000329_790 |
0.1514947751 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 4 |
Hb_003599_020 |
0.158396224 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 5 |
Hb_083493_010 |
0.1708766865 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_008021_010 |
0.1743881496 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 7 |
Hb_000366_050 |
0.1760407467 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000007_440 |
0.176411412 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_071072_010 |
0.1800666156 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_001522_040 |
0.1818291308 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_172676_010 |
0.1821026878 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 12 |
Hb_001158_120 |
0.1844265495 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000936_090 |
0.1849435834 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X2 [Jatropha curcas] |
| 14 |
Hb_005539_100 |
0.1863657854 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 15 |
Hb_002874_170 |
0.1877071731 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_086639_050 |
0.1885550999 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003430_070 |
0.1885848656 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 18 |
Hb_001688_070 |
0.1889704497 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 19 |
Hb_000015_150 |
0.1893697261 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 20 |
Hb_000062_490 |
0.1907593346 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |