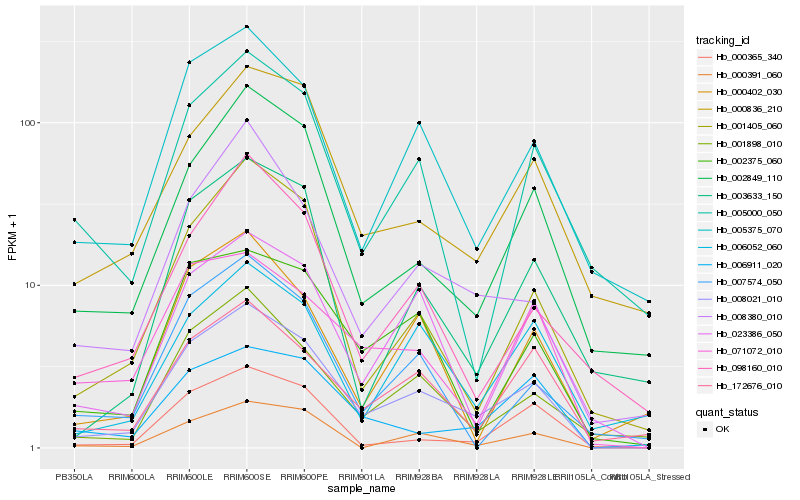
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008021_010 |
0.0 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 2 |
Hb_005375_070 |
0.1350188053 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 3 |
Hb_001898_010 |
0.1424511063 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 4 |
Hb_002375_060 |
0.1471060851 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 5 |
Hb_002849_110 |
0.1502660443 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 6 |
Hb_023386_050 |
0.1517069823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_172676_010 |
0.1536921223 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 8 |
Hb_001405_060 |
0.1557634063 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 9 |
Hb_008380_010 |
0.1560153291 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 10 |
Hb_000836_210 |
0.156303977 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 11 |
Hb_000391_060 |
0.1568564723 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 12 |
Hb_000365_340 |
0.1673253178 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 13 |
Hb_003633_150 |
0.1681534144 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 14 |
Hb_098160_010 |
0.1684992038 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_071072_010 |
0.168563544 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_007574_050 |
0.1705133954 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 17 |
Hb_005000_050 |
0.1710437839 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 18 |
Hb_000402_030 |
0.1714748456 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 19 |
Hb_006911_020 |
0.1715192958 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_006052_060 |
0.1722700764 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |