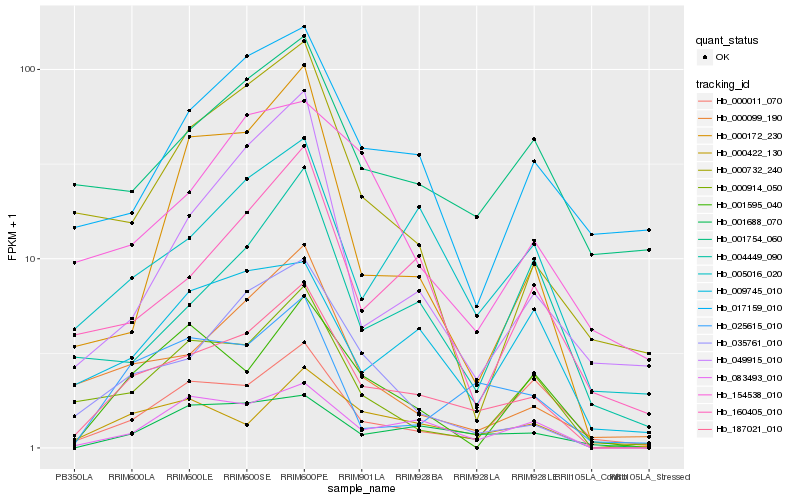
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187021_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_083493_010 |
0.1647932791 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_035761_010 |
0.1725793152 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 4 |
Hb_000422_130 |
0.1850387905 |
- |
- |
hypothetical protein JCGZ_15217 [Jatropha curcas] |
| 5 |
Hb_001688_070 |
0.1853730911 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 6 |
Hb_000099_190 |
0.1866457876 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 7 |
Hb_005016_020 |
0.1882236577 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000914_050 |
0.1887993246 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 9 |
Hb_025615_010 |
0.1914291502 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 10 |
Hb_160405_010 |
0.1952120605 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 11 |
Hb_001754_060 |
0.2086565571 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000172_230 |
0.2110384936 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_000732_240 |
0.2118145431 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 14 |
Hb_009745_010 |
0.2149727428 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 15 |
Hb_017159_010 |
0.2169848394 |
- |
- |
- |
| 16 |
Hb_000011_070 |
0.2170271929 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 17 |
Hb_001595_040 |
0.2198061464 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 18 |
Hb_004449_090 |
0.2199441697 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 19 |
Hb_154538_010 |
0.2209244004 |
- |
- |
- |
| 20 |
Hb_049915_010 |
0.2218740772 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |