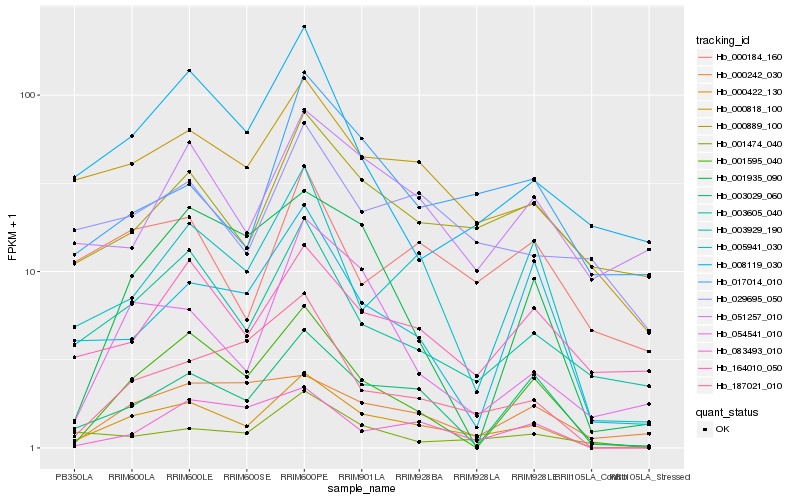
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_15217 [Jatropha curcas] |
| 2 |
Hb_000818_100 |
0.1715443743 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 3 |
Hb_003029_060 |
0.1755671401 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003605_040 |
0.1792194169 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_187021_010 |
0.1850387905 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_003929_190 |
0.1878134491 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 7 |
Hb_001595_040 |
0.1949095642 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 8 |
Hb_000889_100 |
0.2032731181 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 9 |
Hb_054541_010 |
0.2059469723 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 10 |
Hb_083493_010 |
0.2075126015 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 11 |
Hb_164010_050 |
0.2084984867 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 12 |
Hb_001474_040 |
0.2102730023 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 13 |
Hb_051257_010 |
0.2148114013 |
- |
- |
- |
| 14 |
Hb_008119_030 |
0.2155439966 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 15 |
Hb_005941_030 |
0.2158076277 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_017014_010 |
0.2166062687 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_029695_050 |
0.2205655983 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 1 [Jatropha curcas] |
| 18 |
Hb_000242_030 |
0.2238024355 |
- |
- |
PREDICTED: uncharacterized protein LOC102608003 [Citrus sinensis] |
| 19 |
Hb_000184_160 |
0.2238437715 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 20 |
Hb_001935_090 |
0.2245006474 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |