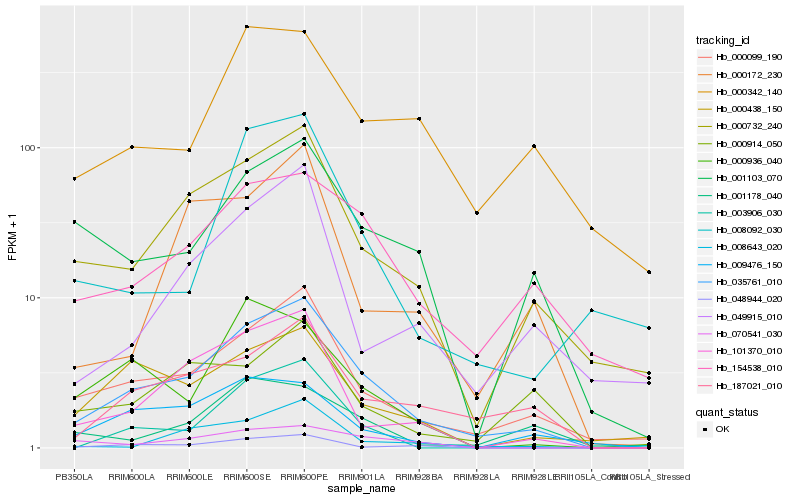
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035761_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 2 |
Hb_000099_190 |
0.1334872869 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 3 |
Hb_000732_240 |
0.1566272923 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 4 |
Hb_187021_010 |
0.1725793152 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_101370_010 |
0.1850998342 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_154538_010 |
0.1854387162 |
- |
- |
- |
| 7 |
Hb_048944_020 |
0.2003594967 |
- |
- |
hypothetical protein POPTR_0014s002002g, partial [Populus trichocarpa] |
| 8 |
Hb_001103_070 |
0.2035804232 |
- |
- |
Patellin-3, putative [Ricinus communis] |
| 9 |
Hb_000438_150 |
0.2073016674 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 10 |
Hb_009476_150 |
0.2099779094 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 11 |
Hb_003906_030 |
0.2110497334 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_049915_010 |
0.2154067225 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 13 |
Hb_000342_140 |
0.2159704667 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 14 |
Hb_008092_030 |
0.2185269349 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 15 |
Hb_008643_020 |
0.2192069283 |
- |
- |
Cf-4/9 disease resistance-like family protein [Populus trichocarpa] |
| 16 |
Hb_070541_030 |
0.2196252947 |
- |
- |
- |
| 17 |
Hb_000172_230 |
0.2232331126 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_001178_040 |
0.2249664492 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 19 |
Hb_000936_040 |
0.2253146871 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 20 |
Hb_000914_050 |
0.2255544065 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |