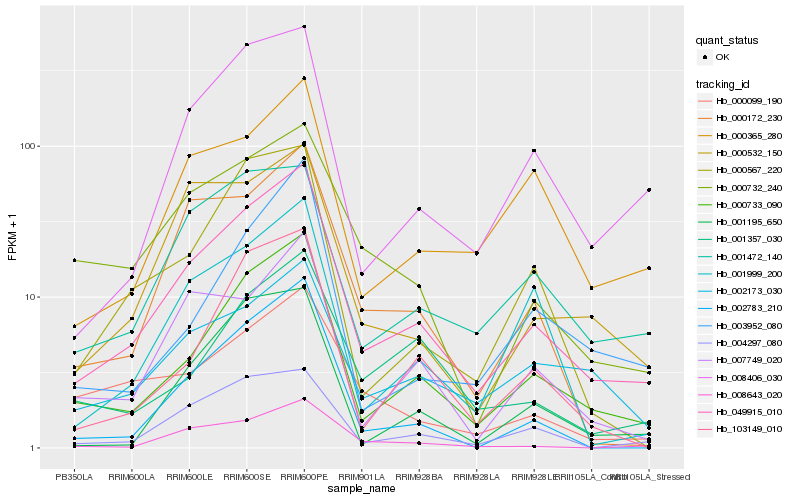
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049915_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 2 |
Hb_000733_090 |
0.0934513085 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 3 |
Hb_008406_030 |
0.1423460225 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 4 |
Hb_004297_080 |
0.1465849306 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_008643_020 |
0.1543695845 |
- |
- |
Cf-4/9 disease resistance-like family protein [Populus trichocarpa] |
| 6 |
Hb_000172_230 |
0.1556541832 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_002783_210 |
0.1564428755 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 8 |
Hb_007749_020 |
0.1568139158 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_002173_030 |
0.1576448116 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 10 |
Hb_001472_140 |
0.158032405 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 11 |
Hb_103149_010 |
0.1585079331 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 12 |
Hb_000732_240 |
0.1622303976 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 13 |
Hb_000532_150 |
0.1638234313 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 14 |
Hb_001999_200 |
0.1661580597 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000365_280 |
0.1667576001 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 16 |
Hb_000567_220 |
0.1670357173 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 17 |
Hb_001195_650 |
0.1693398078 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001357_030 |
0.1721478029 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 19 |
Hb_000099_190 |
0.1733613244 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 20 |
Hb_003952_080 |
0.175729123 |
- |
- |
zinc finger protein, putative [Ricinus communis] |