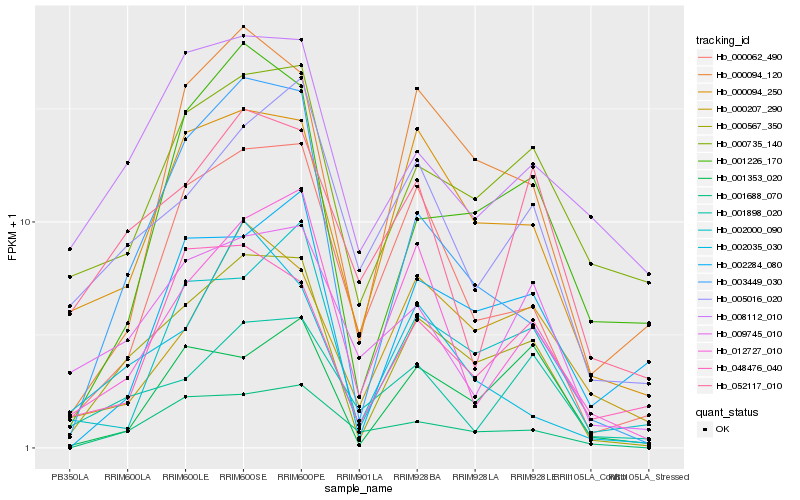
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_350 |
0.0 |
- |
- |
PREDICTED: probable acyl-activating enzyme 6 [Jatropha curcas] |
| 2 |
Hb_000094_250 |
0.1535145063 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 3 |
Hb_003449_030 |
0.1771663888 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 3, chloroplastic-like [Camelina sativa] |
| 4 |
Hb_000062_490 |
0.1776103288 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000094_120 |
0.1785822314 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 6 |
Hb_000735_140 |
0.1795100077 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 7 |
Hb_001688_070 |
0.1807798903 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 8 |
Hb_012727_010 |
0.1844135064 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 9 |
Hb_005016_020 |
0.1850973204 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_002000_090 |
0.1878144541 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 11 |
Hb_009745_010 |
0.1918197949 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 12 |
Hb_001226_170 |
0.1923765781 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 13 |
Hb_001353_020 |
0.1986133016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_048476_040 |
0.1993339476 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 15 |
Hb_008112_010 |
0.1998988449 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 16 |
Hb_002284_080 |
0.1998995129 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001898_020 |
0.2000158688 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 18 |
Hb_000207_290 |
0.2017682482 |
- |
- |
hypothetical protein JCGZ_03920 [Jatropha curcas] |
| 19 |
Hb_002035_030 |
0.2035894104 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |
| 20 |
Hb_052117_010 |
0.2037144543 |
- |
- |
kinase, putative [Ricinus communis] |