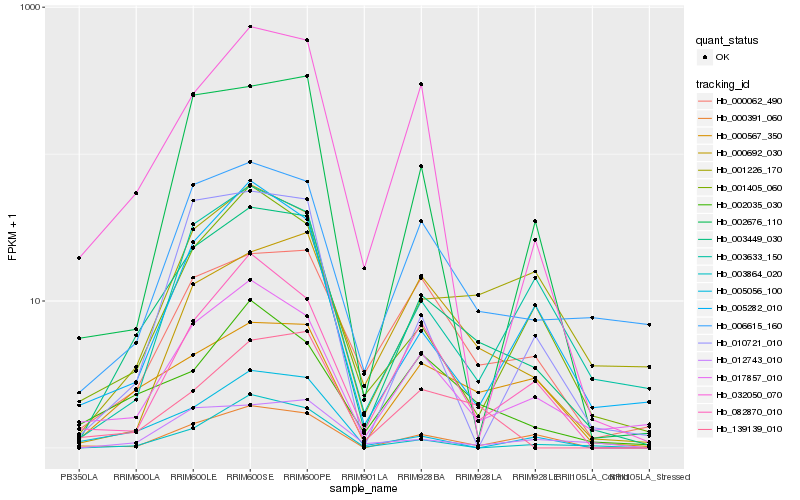
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003449_030 |
0.0 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 3, chloroplastic-like [Camelina sativa] |
| 2 |
Hb_017857_010 |
0.1669495448 |
- |
- |
- |
| 3 |
Hb_006615_160 |
0.1764340612 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 4 |
Hb_000567_350 |
0.1771663888 |
- |
- |
PREDICTED: probable acyl-activating enzyme 6 [Jatropha curcas] |
| 5 |
Hb_012743_010 |
0.1818496145 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 6 |
Hb_000062_490 |
0.1839962023 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 7 |
Hb_032050_070 |
0.1845523821 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 8 |
Hb_139139_010 |
0.1878596335 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000391_060 |
0.1886138011 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 10 |
Hb_001405_060 |
0.1905182389 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 11 |
Hb_005056_100 |
0.1921021356 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 12 |
Hb_002676_110 |
0.193084469 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 2-like [Gossypium raimondii] |
| 13 |
Hb_000692_030 |
0.1939727693 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 14 |
Hb_001226_170 |
0.1942097988 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 15 |
Hb_082870_010 |
0.1947998034 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_002035_030 |
0.1959231041 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |
| 17 |
Hb_003864_020 |
0.1960967747 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_005282_010 |
0.1963490025 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 19 |
Hb_010721_010 |
0.1964014118 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 20 |
Hb_003633_150 |
0.1968173515 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |