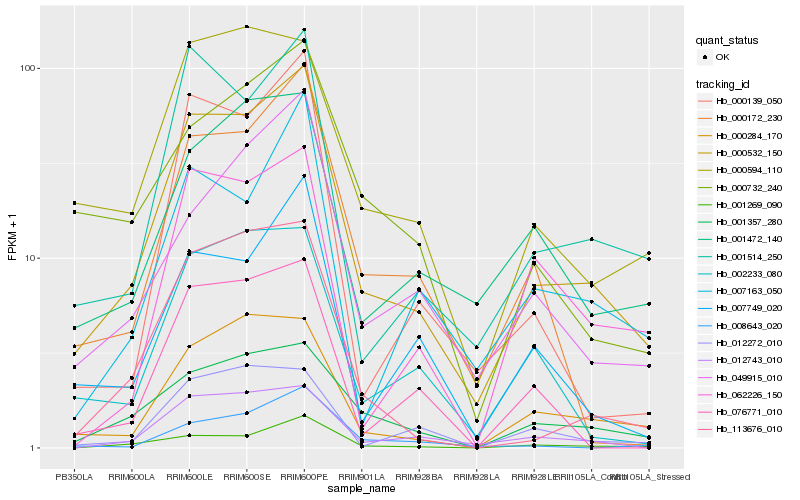
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000532_150 |
0.0 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 2 |
Hb_001514_250 |
0.1347153499 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 3 |
Hb_001269_090 |
0.1430331617 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 4 |
Hb_012743_010 |
0.1499946896 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 5 |
Hb_000284_170 |
0.150540617 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_113676_010 |
0.1531125743 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000172_230 |
0.1532608139 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_001357_280 |
0.1570187393 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 9 |
Hb_049915_010 |
0.1638234313 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 10 |
Hb_007749_020 |
0.1649000483 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 11 |
Hb_000139_050 |
0.166800326 |
- |
- |
PREDICTED: uncharacterized protein LOC105631970 [Jatropha curcas] |
| 12 |
Hb_001472_140 |
0.1676705303 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 13 |
Hb_000594_110 |
0.1699088384 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 14 |
Hb_002233_080 |
0.1705700337 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 15 |
Hb_007163_050 |
0.1751276864 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 16 |
Hb_000732_240 |
0.1782374274 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 17 |
Hb_012272_010 |
0.1807707279 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 18 |
Hb_076771_010 |
0.1818293613 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_008643_020 |
0.1840082024 |
- |
- |
Cf-4/9 disease resistance-like family protein [Populus trichocarpa] |
| 20 |
Hb_062226_150 |
0.1848342147 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |