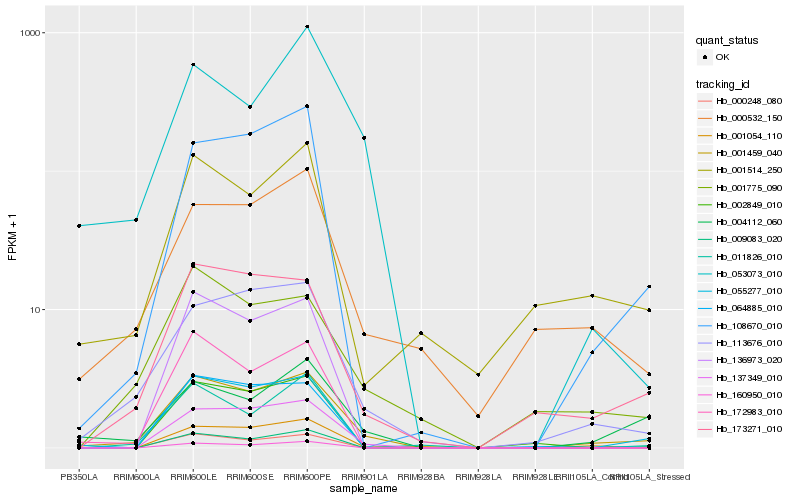
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001459_040 |
0.0 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 2 |
Hb_113676_010 |
0.1425324652 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_173271_010 |
0.1528706058 |
- |
- |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 4 |
Hb_137349_010 |
0.1740690374 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 5 |
Hb_055277_010 |
0.1742240752 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 6 |
Hb_108670_010 |
0.1761964413 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 7 |
Hb_004112_060 |
0.1833653267 |
- |
- |
hypothetical protein POPTR_0017s01950g, partial [Populus trichocarpa] |
| 8 |
Hb_001054_110 |
0.1863906664 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 9 |
Hb_001775_090 |
0.1865914769 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 10 |
Hb_064885_010 |
0.188020238 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 11 |
Hb_000532_150 |
0.1896490877 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 12 |
Hb_001514_250 |
0.1946907303 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 13 |
Hb_172983_010 |
0.195963772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_053073_010 |
0.1967404753 |
- |
- |
- |
| 15 |
Hb_136973_020 |
0.20578464 |
- |
- |
- |
| 16 |
Hb_000248_080 |
0.206604177 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 17 |
Hb_002849_010 |
0.2072386139 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 18 |
Hb_011826_010 |
0.2076512609 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 19 |
Hb_009083_020 |
0.2089542339 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 20 |
Hb_160950_010 |
0.2102008792 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |