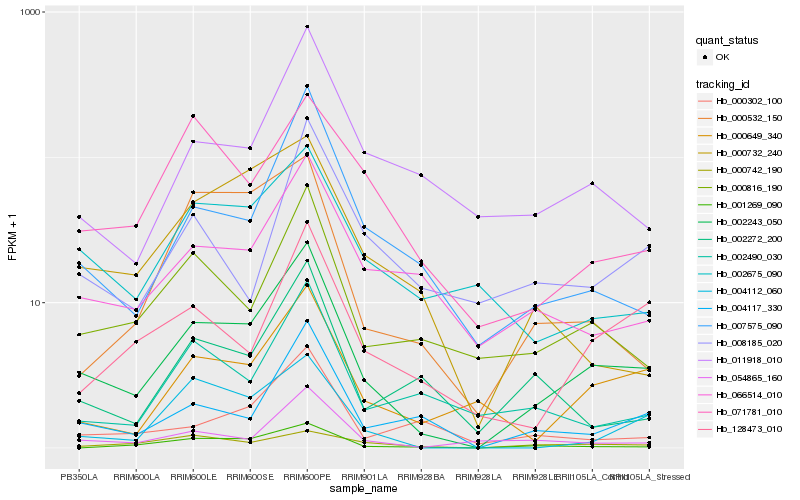
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002243_050 |
0.0 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000816_190 |
0.2009642906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004117_330 |
0.2031894507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000649_340 |
0.2033545272 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_007575_090 |
0.2050215645 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 6 |
Hb_054865_160 |
0.2103049565 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 7 |
Hb_001269_090 |
0.2153791885 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 8 |
Hb_066514_010 |
0.2160191398 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 9 |
Hb_002675_090 |
0.2184426746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004112_060 |
0.2188395076 |
- |
- |
hypothetical protein POPTR_0017s01950g, partial [Populus trichocarpa] |
| 11 |
Hb_128473_010 |
0.2189109181 |
- |
- |
- |
| 12 |
Hb_011918_010 |
0.2217939129 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 13 |
Hb_000532_150 |
0.2352816204 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 14 |
Hb_000742_190 |
0.2357077395 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_002490_030 |
0.2357085703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002272_200 |
0.2375786598 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008185_020 |
0.2380996599 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 18 |
Hb_000732_240 |
0.2382796583 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 19 |
Hb_000302_100 |
0.2387057474 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 20 |
Hb_071781_010 |
0.239147966 |
- |
- |
hypothetical protein POPTR_0010s13190g [Populus trichocarpa] |