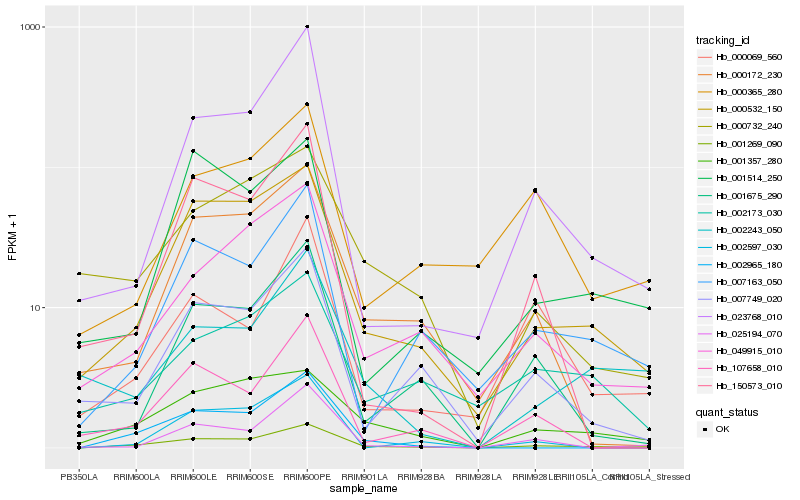
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_090 |
0.0 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 2 |
Hb_000532_150 |
0.1430331617 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 3 |
Hb_007163_050 |
0.1703152678 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 4 |
Hb_049915_010 |
0.1758920137 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 5 |
Hb_023768_010 |
0.183765509 |
- |
- |
- |
| 6 |
Hb_007749_020 |
0.192386464 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_001357_280 |
0.1924988272 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 8 |
Hb_001675_290 |
0.1925160489 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000172_230 |
0.1994906794 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_025194_070 |
0.2015367031 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 11 |
Hb_002965_180 |
0.2018897083 |
- |
- |
AMSH-like ubiquitin thioesterase 3 [Morus notabilis] |
| 12 |
Hb_107658_010 |
0.2097676463 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_002597_030 |
0.2143336896 |
- |
- |
- |
| 14 |
Hb_001514_250 |
0.2146557424 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 15 |
Hb_002243_050 |
0.2153791885 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000365_280 |
0.2156234899 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 17 |
Hb_000069_560 |
0.2161613371 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 18 |
Hb_002173_030 |
0.2164497312 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 19 |
Hb_000732_240 |
0.216721959 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 20 |
Hb_150573_010 |
0.218661418 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |