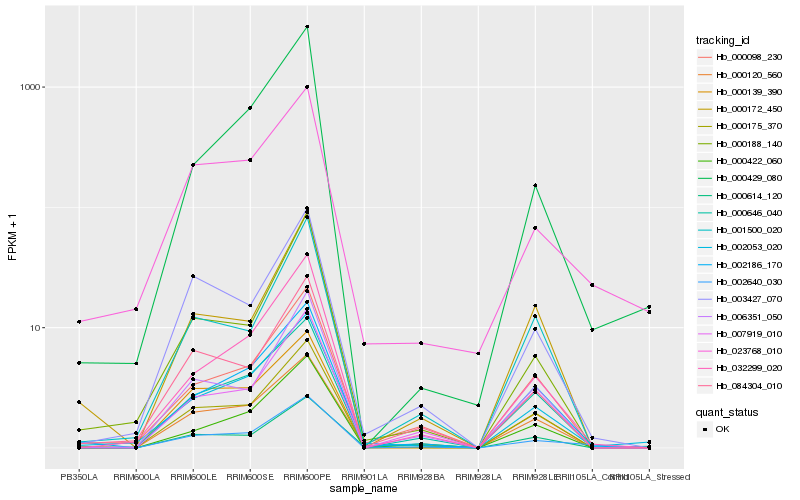
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002640_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s05270g [Populus trichocarpa] |
| 2 |
Hb_002186_170 |
0.1267111744 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 3 |
Hb_023768_010 |
0.1306127443 |
- |
- |
- |
| 4 |
Hb_000175_370 |
0.1346492598 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 5 |
Hb_032299_020 |
0.1375089596 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 6 |
Hb_084304_010 |
0.1405586451 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 7 |
Hb_003427_070 |
0.1428882352 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 8 |
Hb_002053_020 |
0.1431346448 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 9 |
Hb_000120_560 |
0.1457247891 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000098_230 |
0.147437557 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |
| 11 |
Hb_006351_050 |
0.1481121826 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 12 |
Hb_000422_060 |
0.149251827 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 13 |
Hb_007919_010 |
0.1498458824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000646_040 |
0.1536270166 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 15 |
Hb_000172_450 |
0.154049377 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 16 |
Hb_000139_390 |
0.1542780279 |
- |
- |
- |
| 17 |
Hb_000188_140 |
0.1560884761 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 18 |
Hb_000429_080 |
0.1574463293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001500_020 |
0.1575034888 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000614_120 |
0.1582820873 |
- |
- |
hypothetical protein JCGZ_17923 [Jatropha curcas] |