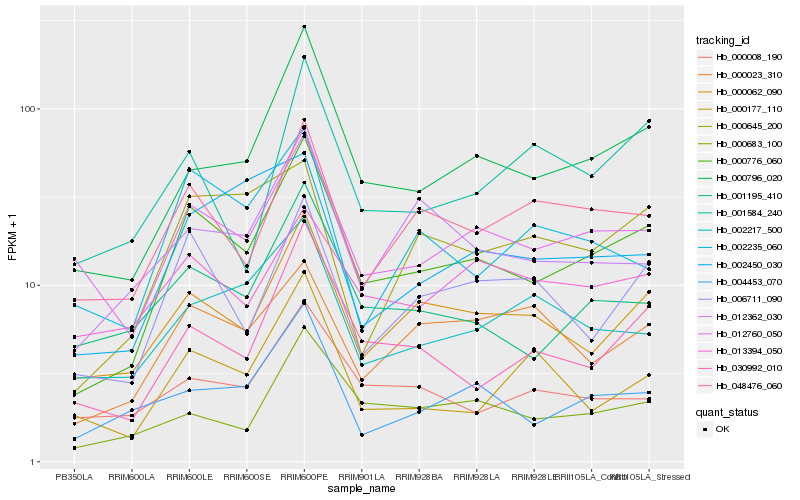
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000776_060 |
0.0 |
- |
- |
histone H4 [Zea mays] |
| 2 |
Hb_000796_020 |
0.121491399 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 3 |
Hb_000062_090 |
0.1357744299 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 4 |
Hb_030992_010 |
0.1420676177 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_012760_050 |
0.1453449982 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 6 |
Hb_001195_410 |
0.1482983532 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 7 |
Hb_006711_090 |
0.1552039112 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 8 |
Hb_048476_060 |
0.1566835991 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_004453_070 |
0.1584714481 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000683_100 |
0.159501064 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 11 |
Hb_012362_030 |
0.1722753755 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 12 |
Hb_000008_190 |
0.1756185131 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002217_500 |
0.1764890322 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 14 |
Hb_002235_060 |
0.1783387942 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_000023_310 |
0.1785521607 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 16 |
Hb_001584_240 |
0.1803208795 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 17 |
Hb_000645_200 |
0.1806177155 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 18 |
Hb_002450_030 |
0.1812475616 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 19 |
Hb_000177_110 |
0.1819987024 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 20 |
Hb_013394_050 |
0.1833639544 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |