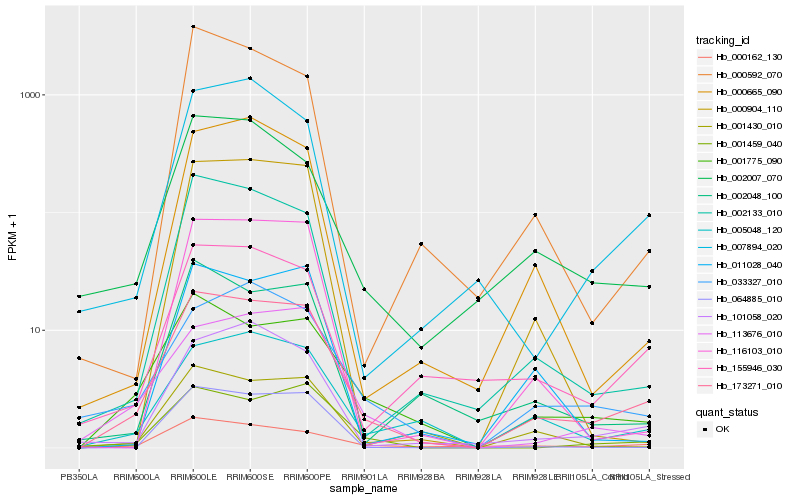
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173271_010 |
0.0 |
- |
- |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 2 |
Hb_001775_090 |
0.1422684787 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 3 |
Hb_000162_130 |
0.1501330178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005048_120 |
0.1521410528 |
- |
- |
- |
| 5 |
Hb_001459_040 |
0.1528706058 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 6 |
Hb_002133_010 |
0.1567645618 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 7 |
Hb_064885_010 |
0.1582907871 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 8 |
Hb_155946_030 |
0.1596459267 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 9 |
Hb_101058_020 |
0.1603588978 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 10 |
Hb_113676_010 |
0.1639471574 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 11 |
Hb_002007_070 |
0.1645347862 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 12 |
Hb_033327_010 |
0.1697484207 |
- |
- |
SYM8 [Pisum sativum] |
| 13 |
Hb_000665_090 |
0.1736620362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_116103_010 |
0.1741448331 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 15 |
Hb_002048_100 |
0.1746160487 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 16 |
Hb_007894_020 |
0.1754116147 |
- |
- |
protein translation factor SUI1 homolog 2 [Jatropha curcas] |
| 17 |
Hb_001430_010 |
0.1794531147 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011028_040 |
0.1826209826 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 19 |
Hb_000904_110 |
0.185352163 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF109-like [Eucalyptus grandis] |
| 20 |
Hb_000592_070 |
0.1885806864 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |