| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
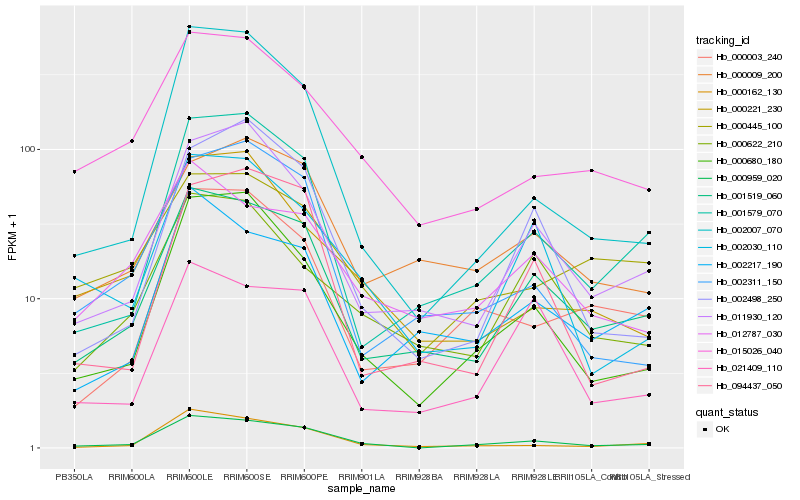
Hb_000959_020 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_000680_180 |
0.1267008102 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 3 |
Hb_001519_060 |
0.1306206554 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000162_130 |
0.1431189339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001579_070 |
0.1538284993 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 6 |
Hb_002007_070 |
0.1562129283 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 7 |
Hb_015026_040 |
0.1567142944 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 8 |
Hb_002030_110 |
0.1623575124 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 9 |
Hb_011930_120 |
0.1635499818 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000003_240 |
0.1680975892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002498_250 |
0.172503231 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000221_230 |
0.1767066432 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 13 |
Hb_002311_150 |
0.1782520763 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 14 |
Hb_094437_050 |
0.1793193011 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 15 |
Hb_012787_030 |
0.1840716954 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 16 |
Hb_000622_210 |
0.1848228728 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_021409_110 |
0.1884580272 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_002217_190 |
0.1902006662 |
- |
- |
invertase [Hevea brasiliensis] |
| 19 |
Hb_000445_100 |
0.1904782138 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 20 |
Hb_000009_200 |
0.1913195155 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |