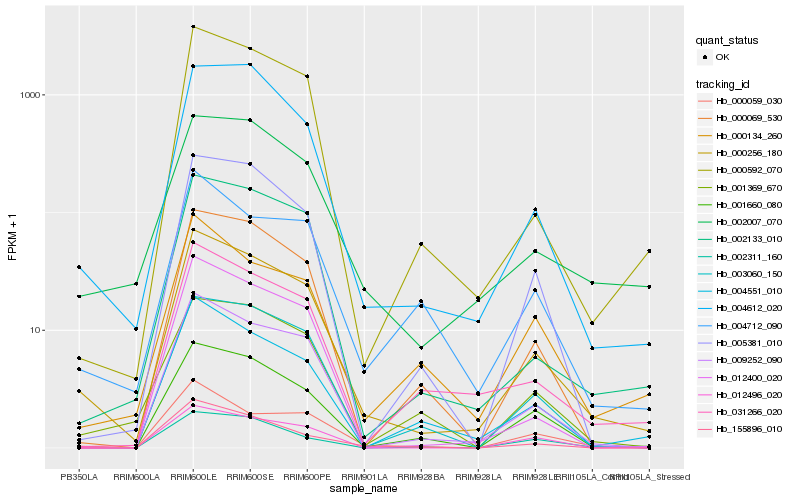
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637589 [Jatropha curcas] |
| 2 |
Hb_009252_090 |
0.1211477033 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_004612_020 |
0.1230183873 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 4 |
Hb_000592_070 |
0.1251330405 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 5 |
Hb_002133_010 |
0.1314581318 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 6 |
Hb_001660_080 |
0.133137391 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 7 |
Hb_002007_070 |
0.1343666635 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 8 |
Hb_000059_030 |
0.1351492314 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 9 |
Hb_000069_530 |
0.1372891199 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 10 |
Hb_031266_020 |
0.1379304705 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005381_010 |
0.1403640884 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003060_150 |
0.1455513143 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_012400_020 |
0.1460732316 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 14 |
Hb_004551_010 |
0.1477843676 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 15 |
Hb_155896_010 |
0.1490318959 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 16 |
Hb_004712_090 |
0.1498331535 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_001369_670 |
0.1503092582 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 18 |
Hb_000134_260 |
0.1518256576 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_002311_160 |
0.1525857498 |
transcription factor |
TF Family: ARR-B |
sensor histidine kinase, putative [Ricinus communis] |
| 20 |
Hb_012496_020 |
0.1537191955 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |