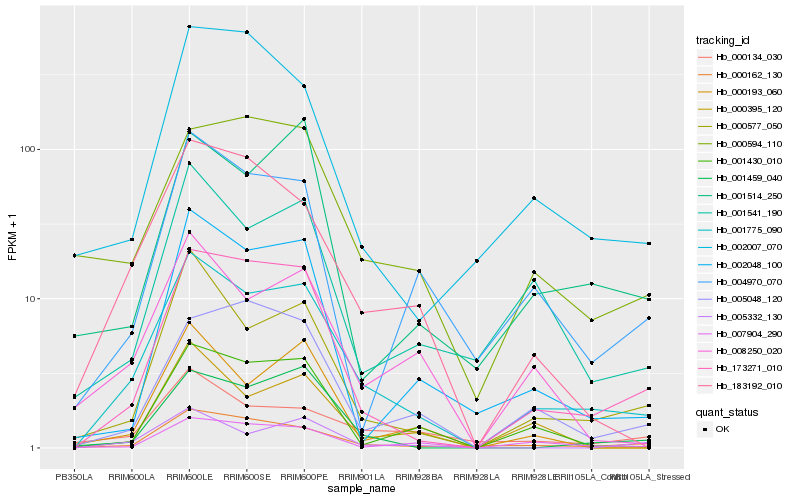
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001775_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 2 |
Hb_173271_010 |
0.1422684787 |
- |
- |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 3 |
Hb_000577_050 |
0.1534083274 |
- |
- |
- |
| 4 |
Hb_000162_130 |
0.1614731125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_183192_010 |
0.1848064686 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 6 |
Hb_001459_040 |
0.1865914769 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 7 |
Hb_001541_190 |
0.1871227432 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 8 |
Hb_008250_020 |
0.1878314151 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001430_010 |
0.1878460201 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002048_100 |
0.1915914743 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Jatropha curcas] |
| 11 |
Hb_005332_130 |
0.1918025371 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 12 |
Hb_004970_070 |
0.1955321562 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 13 |
Hb_007904_290 |
0.1961156877 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 14 |
Hb_000134_030 |
0.2007681297 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 15 |
Hb_000395_120 |
0.2015015594 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 16 |
Hb_000193_060 |
0.2052493518 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 17 |
Hb_000594_110 |
0.207619384 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 18 |
Hb_002007_070 |
0.2093949433 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 19 |
Hb_001514_250 |
0.2094185632 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 20 |
Hb_005048_120 |
0.2095140204 |
- |
- |
- |