| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
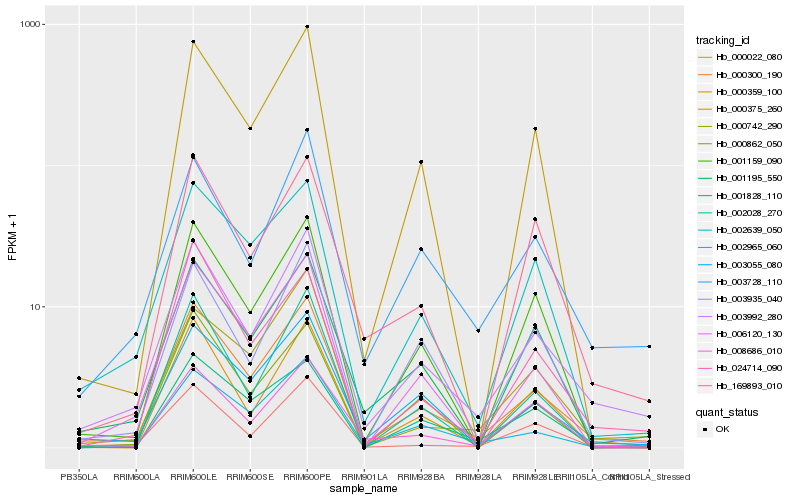
Hb_000300_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 2 |
Hb_003992_280 |
0.0626902012 |
- |
- |
PREDICTED: uncharacterized protein LOC105117987 [Populus euphratica] |
| 3 |
Hb_003055_080 |
0.0966746345 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002965_060 |
0.0975365118 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 5 |
Hb_006120_130 |
0.1145437678 |
- |
- |
- |
| 6 |
Hb_002639_050 |
0.1157230594 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 7 |
Hb_000375_260 |
0.1177645141 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 8 |
Hb_001195_550 |
0.1190148442 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002028_270 |
0.1207683708 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_001159_090 |
0.1218727219 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 11 |
Hb_001828_110 |
0.1256141342 |
- |
- |
PREDICTED: lysine histidine transporter 1-like [Tarenaya hassleriana] |
| 12 |
Hb_003728_110 |
0.127953588 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 13 |
Hb_024714_090 |
0.1302257893 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 14 |
Hb_003935_040 |
0.13305979 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 15 |
Hb_000359_100 |
0.1337793793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000022_080 |
0.1352245484 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 17 |
Hb_008686_010 |
0.1356181055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000862_050 |
0.1371712491 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 19 |
Hb_169893_010 |
0.1388015631 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 20 |
Hb_000742_290 |
0.13907008 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |