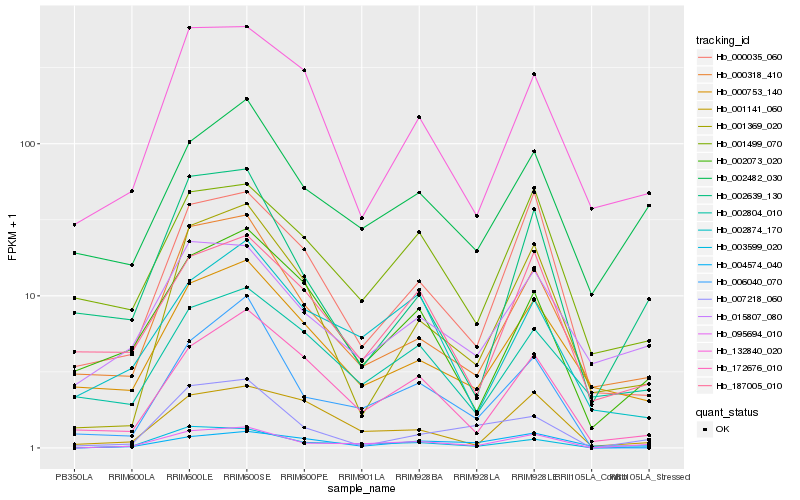
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_095694_010 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Populus euphratica] |
| 2 |
Hb_003599_020 |
0.1372346934 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 3 |
Hb_002482_030 |
0.1780667806 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 4 |
Hb_015807_080 |
0.1821323682 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 5 |
Hb_000318_410 |
0.1856093989 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_000035_060 |
0.1960762952 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 7 |
Hb_172676_010 |
0.2004218372 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 8 |
Hb_002639_130 |
0.202611161 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 9 |
Hb_001499_070 |
0.2029888284 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 10 |
Hb_001369_020 |
0.2031401428 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 11 |
Hb_004574_040 |
0.2051514972 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 12 |
Hb_006040_070 |
0.2054296976 |
- |
- |
orf107b gene product (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_187005_010 |
0.2059254955 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 14 |
Hb_000753_140 |
0.2064357212 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 15 |
Hb_002073_020 |
0.208107097 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 16 |
Hb_001141_060 |
0.2086982041 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_132840_020 |
0.2091694105 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 18 |
Hb_007218_060 |
0.2093541339 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002804_010 |
0.2146181692 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 20 |
Hb_002874_170 |
0.2161491736 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |