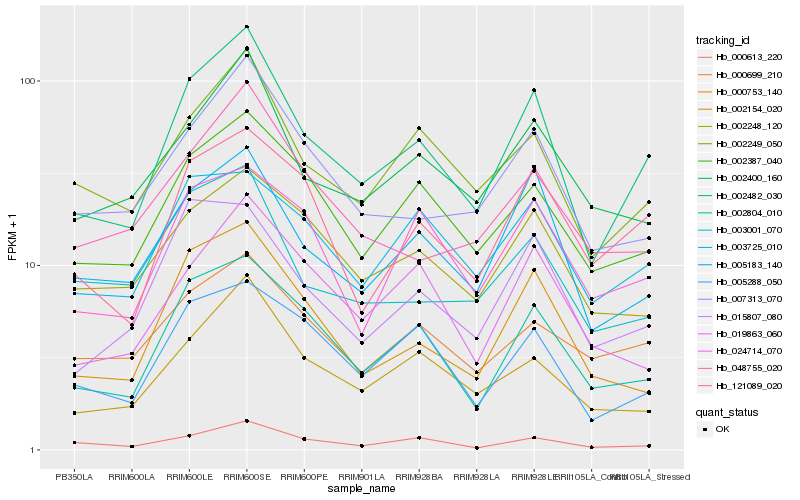
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002482_030 |
0.0 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 2 |
Hb_002804_010 |
0.110784283 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 3 |
Hb_005183_140 |
0.1159745207 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 4 |
Hb_002249_050 |
0.1189007857 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 5 |
Hb_002387_040 |
0.1232954948 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 6 |
Hb_002154_020 |
0.1264137058 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_002400_160 |
0.1266287134 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 8 |
Hb_000753_140 |
0.1277218459 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 9 |
Hb_007313_070 |
0.1282183406 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 10 |
Hb_000613_220 |
0.1283996048 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 11 |
Hb_121089_020 |
0.1297089406 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 12 |
Hb_015807_080 |
0.1341509242 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 13 |
Hb_005288_050 |
0.1368793003 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 14 |
Hb_002248_120 |
0.1444747721 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 15 |
Hb_003725_010 |
0.1455201765 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 16 |
Hb_019863_060 |
0.1460621764 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 17 |
Hb_048755_020 |
0.1461908052 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 18 |
Hb_024714_070 |
0.1462027937 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 19 |
Hb_000699_210 |
0.1474788273 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_003001_070 |
0.1484715973 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |