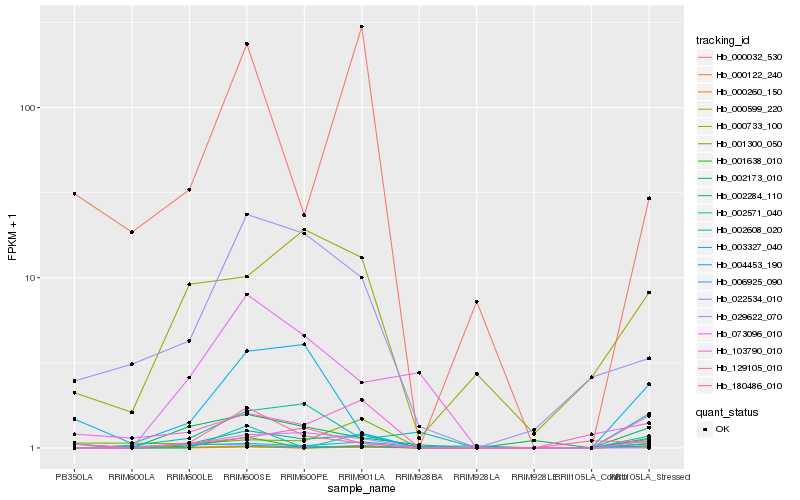
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_110 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP18, chloroplastic isoform X2 [Solanum lycopersicum] |
| 2 |
Hb_000599_220 |
0.1252405735 |
- |
- |
PREDICTED: uncharacterized protein LOC105644022 [Jatropha curcas] |
| 3 |
Hb_000733_100 |
0.3142226383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002173_010 |
0.3203315582 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_006925_090 |
0.3244403082 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 6 |
Hb_022534_010 |
0.3264550889 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 7 |
Hb_103790_010 |
0.3322709528 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 8 |
Hb_000260_150 |
0.3399380154 |
- |
- |
hypothetical protein POPTR_0344s002001g, partial [Populus trichocarpa] |
| 9 |
Hb_029622_070 |
0.3490246806 |
- |
- |
- |
| 10 |
Hb_129105_010 |
0.3498092026 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_001638_010 |
0.3595360747 |
- |
- |
- |
| 12 |
Hb_000032_530 |
0.3599433404 |
- |
- |
- |
| 13 |
Hb_002608_020 |
0.361674754 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 305 [Jatropha curcas] |
| 14 |
Hb_002571_040 |
0.3620833374 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 15 |
Hb_073096_010 |
0.3632782652 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 45-like [Jatropha curcas] |
| 16 |
Hb_180486_010 |
0.3695342192 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_004453_190 |
0.3720436768 |
- |
- |
- |
| 18 |
Hb_003327_040 |
0.3737931983 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 19 |
Hb_000122_240 |
0.3763482429 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 20 |
Hb_001300_050 |
0.3804528578 |
- |
- |
- |