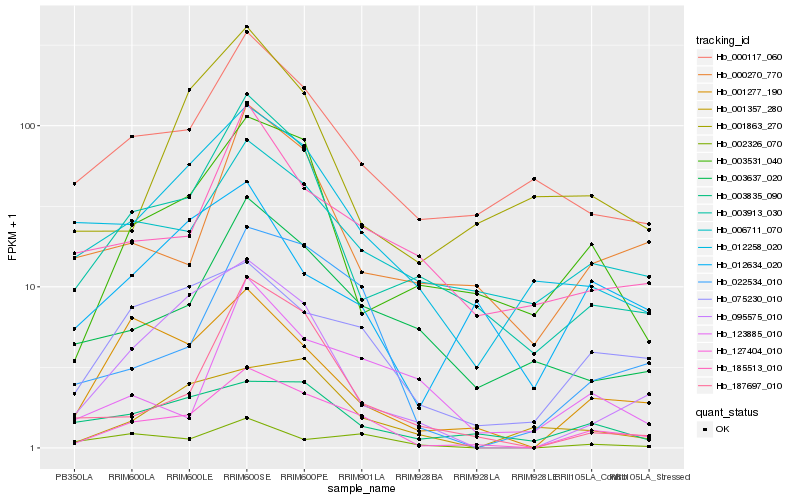
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127404_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X1 [Gossypium raimondii] |
| 2 |
Hb_022534_010 |
0.1775238109 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 3 |
Hb_075230_010 |
0.1984395253 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 4 |
Hb_003913_030 |
0.2128852824 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 5 |
Hb_012258_020 |
0.2175481818 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_001277_190 |
0.2202139458 |
- |
- |
hypothetical protein CICLE_v10005534mg [Citrus clementina] |
| 7 |
Hb_000117_060 |
0.2207627214 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 8 |
Hb_003637_020 |
0.2208944828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003531_040 |
0.2222455209 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_095575_010 |
0.2248545478 |
- |
- |
hypothetical protein JCGZ_19529 [Jatropha curcas] |
| 11 |
Hb_006711_070 |
0.2262401669 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 12 |
Hb_000270_770 |
0.2319589227 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 13 |
Hb_185513_010 |
0.232385965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_187697_010 |
0.2324527877 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 15 |
Hb_003835_090 |
0.2373240308 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 16 |
Hb_002326_070 |
0.2449462811 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001357_280 |
0.2470919222 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 18 |
Hb_123885_010 |
0.2499188629 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 19 |
Hb_012634_020 |
0.2514205802 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001863_270 |
0.2573434455 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |