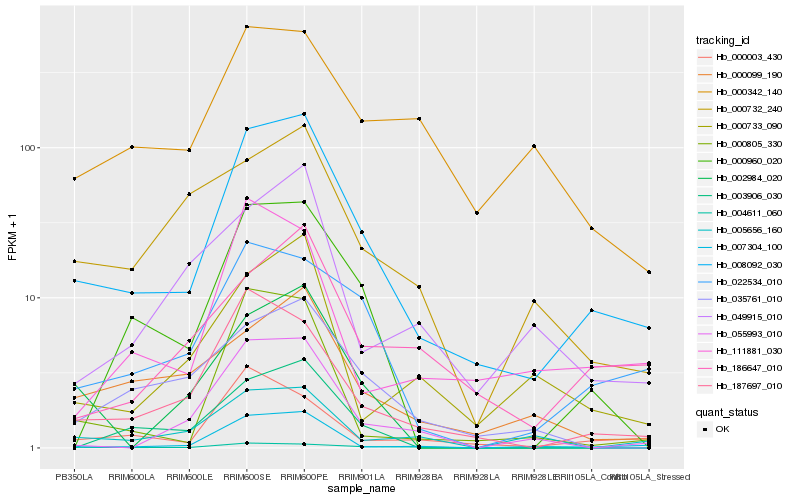
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008092_030 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 2 |
Hb_187697_010 |
0.1825759722 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 3 |
Hb_007304_100 |
0.1866360241 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 4 |
Hb_022534_010 |
0.1916828748 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 5 |
Hb_000960_020 |
0.2104987846 |
- |
- |
- |
| 6 |
Hb_000805_330 |
0.2113348223 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 7 |
Hb_000099_190 |
0.2162974976 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 8 |
Hb_055993_010 |
0.2182194106 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_035761_010 |
0.2185269349 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 10 |
Hb_186647_010 |
0.218924851 |
- |
- |
- |
| 11 |
Hb_000003_430 |
0.2195519881 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_000733_090 |
0.2212660424 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 13 |
Hb_111881_030 |
0.2279817491 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 14 |
Hb_004611_060 |
0.2312452923 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 15 |
Hb_049915_010 |
0.2320607672 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 16 |
Hb_005656_160 |
0.2330983953 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 17 |
Hb_002984_020 |
0.2337305043 |
- |
- |
hypothetical protein EUGRSUZ_J02661 [Eucalyptus grandis] |
| 18 |
Hb_000342_140 |
0.2338961089 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 19 |
Hb_000732_240 |
0.2343862288 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 20 |
Hb_003906_030 |
0.2344040474 |
- |
- |
mads box protein, putative [Ricinus communis] |