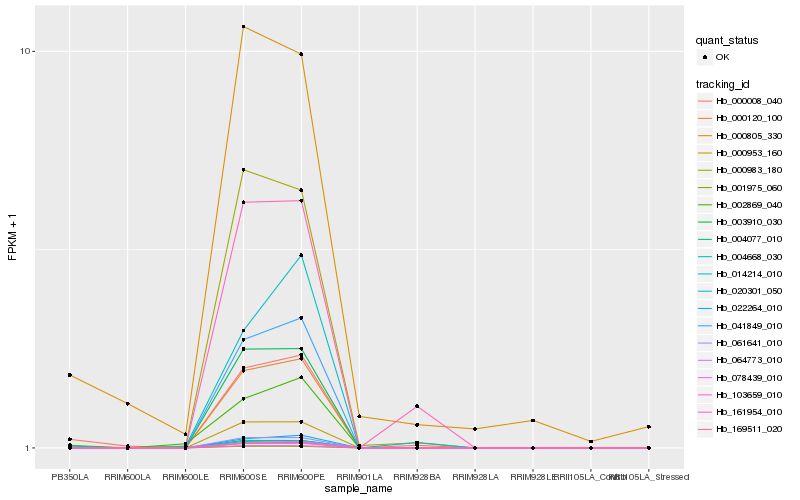
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s01550g [Populus trichocarpa] |
| 2 |
Hb_000805_330 |
0.1592057598 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 3 |
Hb_002869_040 |
0.1677539764 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 4 |
Hb_064773_010 |
0.1688882436 |
- |
- |
PREDICTED: uncharacterized protein LOC103699620, partial [Phoenix dactylifera] |
| 5 |
Hb_000120_100 |
0.1689050497 |
- |
- |
- |
| 6 |
Hb_004668_030 |
0.1691284409 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 7 |
Hb_020301_050 |
0.1691571136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_041849_010 |
0.1693630544 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 9 |
Hb_078439_010 |
0.169363766 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 10 |
Hb_022264_010 |
0.1717775464 |
- |
- |
PREDICTED: uncharacterized protein LOC105795741 [Gossypium raimondii] |
| 11 |
Hb_169511_020 |
0.1724265705 |
- |
- |
PREDICTED: uncharacterized protein LOC105037523, partial [Elaeis guineensis] |
| 12 |
Hb_014214_010 |
0.1726311704 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_003910_030 |
0.17333625 |
- |
- |
PREDICTED: probable disease resistance protein At5g66910 [Populus euphratica] |
| 14 |
Hb_000953_160 |
0.1734261777 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 15 |
Hb_061641_010 |
0.1737716398 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 16 |
Hb_004077_010 |
0.1744056674 |
- |
- |
retinoblastoma-binding protein, putative [Ricinus communis] |
| 17 |
Hb_103659_010 |
0.1746203495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_161954_010 |
0.1746252531 |
- |
- |
hypothetical protein JCGZ_05404 [Jatropha curcas] |
| 19 |
Hb_000983_180 |
0.1756893071 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_001975_060 |
0.1766063871 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |