| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
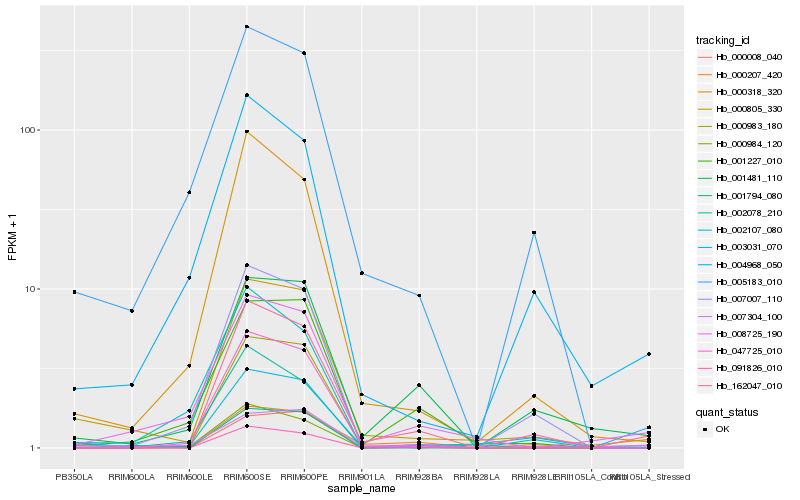
Hb_000805_330 |
0.0 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 2 |
Hb_007304_100 |
0.1551621382 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 3 |
Hb_000008_040 |
0.1592057598 |
- |
- |
hypothetical protein POPTR_0009s01550g [Populus trichocarpa] |
| 4 |
Hb_000318_320 |
0.1658629662 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 5 |
Hb_008725_190 |
0.1726195943 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 6 |
Hb_005183_010 |
0.1736296412 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 7 |
Hb_007007_110 |
0.1808938706 |
- |
- |
hypothetical protein JCGZ_14479 [Jatropha curcas] |
| 8 |
Hb_002078_210 |
0.1822919037 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |
| 9 |
Hb_047725_010 |
0.1833101477 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 10 |
Hb_000983_180 |
0.1849349464 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_001794_080 |
0.1886602385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_162047_010 |
0.1913651087 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 13 |
Hb_001227_010 |
0.1923250764 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 14 |
Hb_000207_420 |
0.1928451144 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 15 |
Hb_004968_050 |
0.1938825126 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 16 |
Hb_003031_070 |
0.1944927413 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 17 |
Hb_001481_110 |
0.198864473 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 18 |
Hb_000984_120 |
0.1995976067 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_002107_080 |
0.2016298806 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 20 |
Hb_091826_010 |
0.2035367316 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |