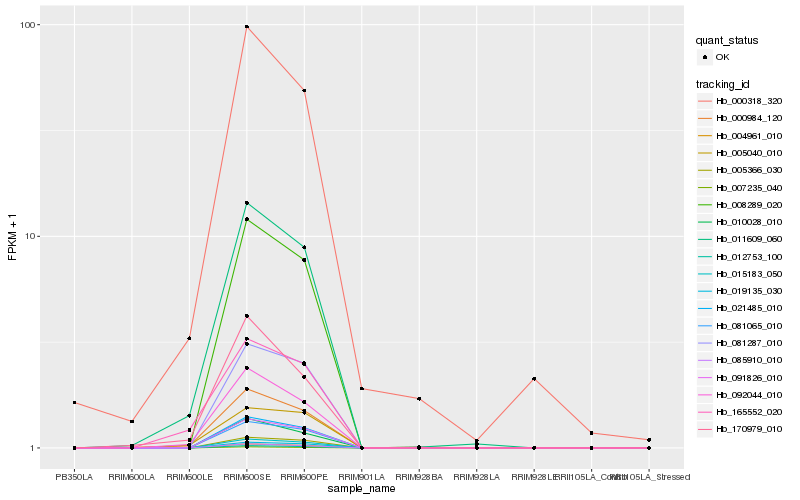
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000984_120 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_011609_060 |
0.0688696202 |
- |
- |
PREDICTED: cytokinin hydroxylase [Jatropha curcas] |
| 3 |
Hb_092044_010 |
0.0857175087 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 4 |
Hb_170979_010 |
0.1023800567 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 5 |
Hb_091826_010 |
0.1052283843 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000318_320 |
0.1056185355 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 7 |
Hb_085910_010 |
0.1173453022 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 8 |
Hb_012753_100 |
0.1175750411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_021485_010 |
0.1187554141 |
- |
- |
Integrase-type DNA-binding superfamily protein, putative [Theobroma cacao] |
| 10 |
Hb_008289_020 |
0.118839515 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 11 |
Hb_004961_010 |
0.1189991146 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 12 |
Hb_015183_050 |
0.1210410854 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 13 |
Hb_010028_010 |
0.1223963753 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 14 |
Hb_081065_010 |
0.1226982623 |
- |
- |
hypothetical protein PHAVU_001G084100g, partial [Phaseolus vulgaris] |
| 15 |
Hb_007235_040 |
0.1233999776 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_005040_010 |
0.1237320457 |
- |
- |
hypothetical protein RCOM_1110360 [Ricinus communis] |
| 17 |
Hb_165552_020 |
0.1245897354 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_005366_030 |
0.127485637 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 19 |
Hb_019135_030 |
0.1281296233 |
- |
- |
putative mitogen-activated protein kinase 1 [Populus trichocarpa] |
| 20 |
Hb_081287_010 |
0.1287475512 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Jatropha curcas] |