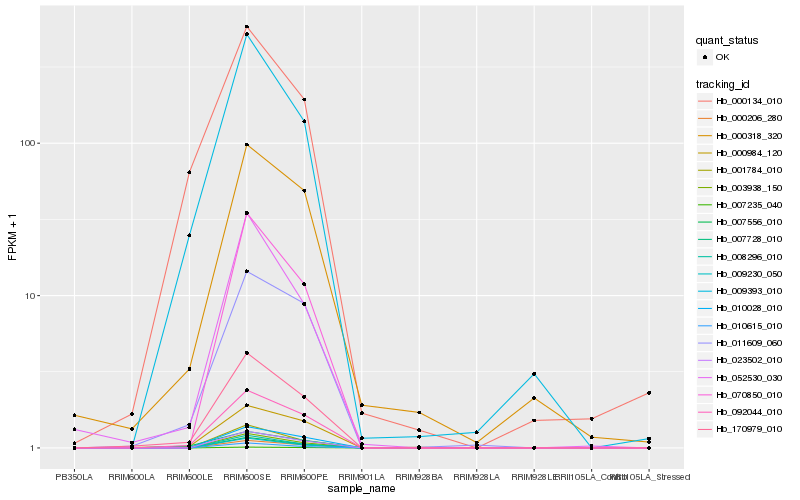
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170979_010 |
0.0 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 2 |
Hb_092044_010 |
0.0731617855 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 3 |
Hb_009393_010 |
0.0986654565 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 4 |
Hb_000984_120 |
0.1023800567 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_011609_060 |
0.1060732849 |
- |
- |
PREDICTED: cytokinin hydroxylase [Jatropha curcas] |
| 6 |
Hb_023502_010 |
0.1157719066 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 7 |
Hb_000206_280 |
0.1161316276 |
- |
- |
Uncharacterized protein TCM_031897 [Theobroma cacao] |
| 8 |
Hb_003938_150 |
0.1166035999 |
- |
- |
hypothetical protein JCGZ_17711 [Jatropha curcas] |
| 9 |
Hb_007556_010 |
0.1168895937 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X2 [Populus euphratica] |
| 10 |
Hb_070850_010 |
0.1170805473 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B-like [Jatropha curcas] |
| 11 |
Hb_052530_030 |
0.1172126848 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 12 |
Hb_007728_010 |
0.1220715531 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 13 |
Hb_007235_040 |
0.1230865528 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_009230_050 |
0.1230969221 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 15 |
Hb_010028_010 |
0.1243302945 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 16 |
Hb_008296_010 |
0.1251189307 |
- |
- |
- |
| 17 |
Hb_001784_010 |
0.1257092257 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 18 |
Hb_010615_010 |
0.1280228364 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 19 |
Hb_000318_320 |
0.1294088159 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 20 |
Hb_000134_010 |
0.1303958654 |
- |
- |
- |