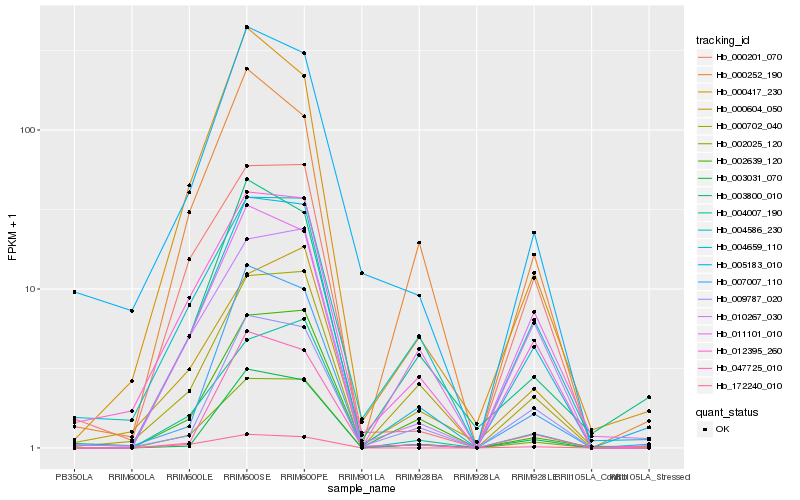
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004659_110 |
0.0 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 2 |
Hb_002025_120 |
0.0561007056 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 3 |
Hb_012395_260 |
0.1089310274 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004007_190 |
0.1102717795 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 5 |
Hb_009787_020 |
0.116911601 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 6 |
Hb_010267_030 |
0.1240939653 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000201_070 |
0.124853949 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 8 |
Hb_172240_010 |
0.1250151793 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 9 |
Hb_002639_120 |
0.1275841184 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_007007_110 |
0.1277601574 |
- |
- |
hypothetical protein JCGZ_14479 [Jatropha curcas] |
| 11 |
Hb_003031_070 |
0.128921969 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 12 |
Hb_000702_040 |
0.1294544093 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 13 |
Hb_047725_010 |
0.1344869853 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 14 |
Hb_005183_010 |
0.1391112445 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 15 |
Hb_000604_050 |
0.1425724098 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 16 |
Hb_000417_230 |
0.1439113433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004586_230 |
0.1468631295 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003800_010 |
0.1468926827 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 19 |
Hb_011101_010 |
0.1490447783 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 20 |
Hb_000252_190 |
0.1517815978 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |