| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
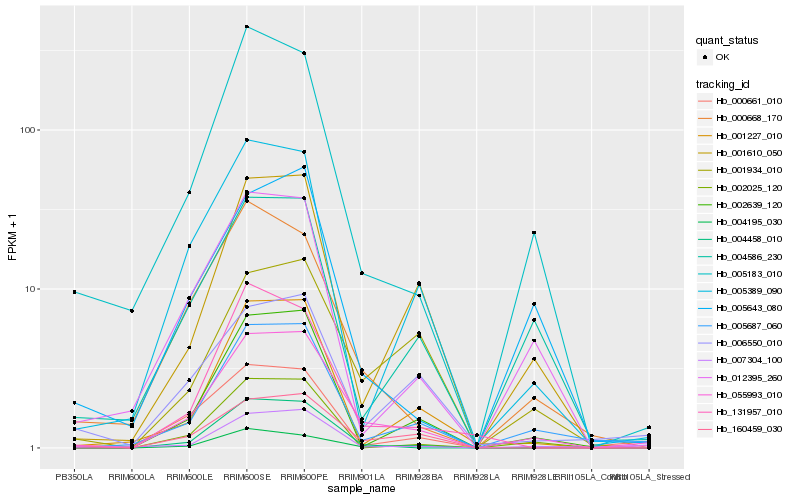
Hb_055993_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_005687_060 |
0.1392950274 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_160459_030 |
0.1559463232 |
- |
- |
PREDICTED: uncharacterized protein LOC105648122 [Jatropha curcas] |
| 4 |
Hb_005183_010 |
0.1607595675 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 5 |
Hb_002639_120 |
0.1679202895 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_007304_100 |
0.1709817572 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 7 |
Hb_004195_030 |
0.1719774967 |
- |
- |
hypothetical protein POPTR_0006s17880g, partial [Populus trichocarpa] |
| 8 |
Hb_001610_050 |
0.1722611904 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 9 |
Hb_004586_230 |
0.1752257093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005389_090 |
0.1764121088 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 11 |
Hb_002025_120 |
0.1774563359 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 12 |
Hb_131957_010 |
0.1779057332 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 13 |
Hb_012395_260 |
0.179668041 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001934_010 |
0.1798115312 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000668_170 |
0.1816532815 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 16 |
Hb_004458_010 |
0.1819479362 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 17 |
Hb_001227_010 |
0.1828908564 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 18 |
Hb_005643_080 |
0.1831454478 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 19 |
Hb_006550_010 |
0.1845670138 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 20 |
Hb_000661_010 |
0.1845993296 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |