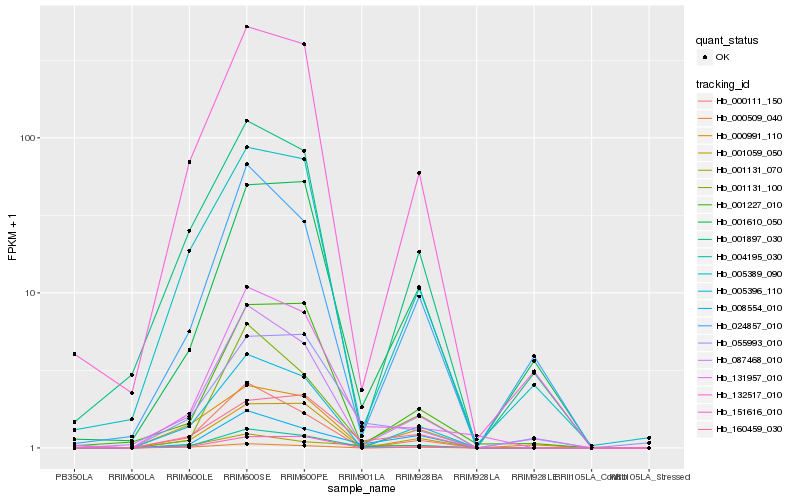
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004195_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s17880g, partial [Populus trichocarpa] |
| 2 |
Hb_131957_010 |
0.1265605229 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_005396_110 |
0.127813551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_132517_010 |
0.13412303 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 5 |
Hb_087468_010 |
0.1373689298 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 6 |
Hb_008554_010 |
0.1401761895 |
- |
- |
- |
| 7 |
Hb_160459_030 |
0.1405746983 |
- |
- |
PREDICTED: uncharacterized protein LOC105648122 [Jatropha curcas] |
| 8 |
Hb_001131_100 |
0.1410205312 |
- |
- |
PREDICTED: uncharacterized protein LOC105638386 [Jatropha curcas] |
| 9 |
Hb_000111_150 |
0.1525995579 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 10 |
Hb_000509_040 |
0.1541786834 |
- |
- |
hypothetical protein VITISV_009716 [Vitis vinifera] |
| 11 |
Hb_001059_050 |
0.1587884112 |
- |
- |
PREDICTED: putative F-box/LRR-repeat protein At3g18150 [Jatropha curcas] |
| 12 |
Hb_151616_010 |
0.1624652241 |
- |
- |
- |
| 13 |
Hb_001131_070 |
0.1667004923 |
- |
- |
PREDICTED: uncharacterized protein LOC105638217 [Jatropha curcas] |
| 14 |
Hb_005389_090 |
0.1686890488 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 15 |
Hb_001897_030 |
0.1691159628 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 16 |
Hb_055993_010 |
0.1719774967 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_001227_010 |
0.1735614511 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 18 |
Hb_024857_010 |
0.1748891305 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 19 |
Hb_000991_110 |
0.1754526651 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 20 |
Hb_001610_050 |
0.1772398259 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |