| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
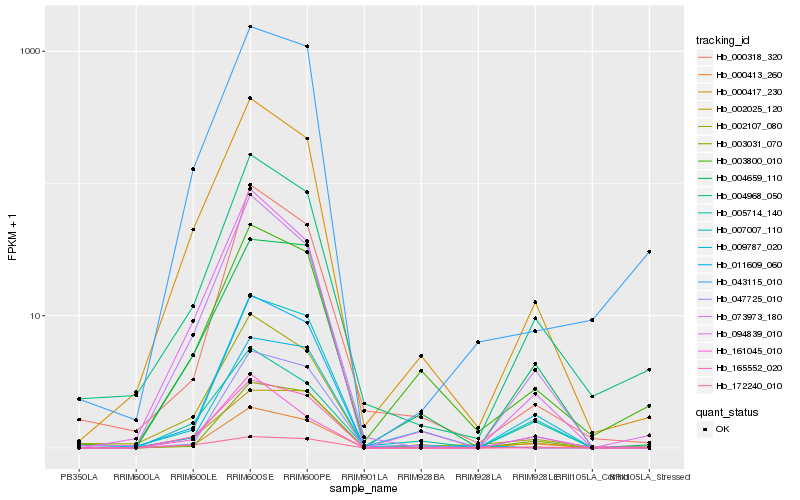
Hb_000413_260 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_007007_110 |
0.1243168415 |
- |
- |
hypothetical protein JCGZ_14479 [Jatropha curcas] |
| 3 |
Hb_047725_010 |
0.1412149164 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 4 |
Hb_073973_180 |
0.1413147568 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 5 |
Hb_000417_230 |
0.1493441187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002107_080 |
0.1505470632 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 7 |
Hb_004659_110 |
0.1542728602 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 8 |
Hb_003031_070 |
0.156925423 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 55-like [Jatropha curcas] |
| 9 |
Hb_004968_050 |
0.1595571239 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 10 |
Hb_011609_060 |
0.1683451453 |
- |
- |
PREDICTED: cytokinin hydroxylase [Jatropha curcas] |
| 11 |
Hb_094839_010 |
0.1691765368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000318_320 |
0.1727239345 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 13 |
Hb_002025_120 |
0.1747160792 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 14 |
Hb_043115_010 |
0.1748283888 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |
| 15 |
Hb_161045_010 |
0.1789379793 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 16 |
Hb_009787_020 |
0.1791950141 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 17 |
Hb_005714_140 |
0.1797162935 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_003800_010 |
0.1816320989 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 19 |
Hb_165552_020 |
0.1821337418 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_172240_010 |
0.1825749776 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |