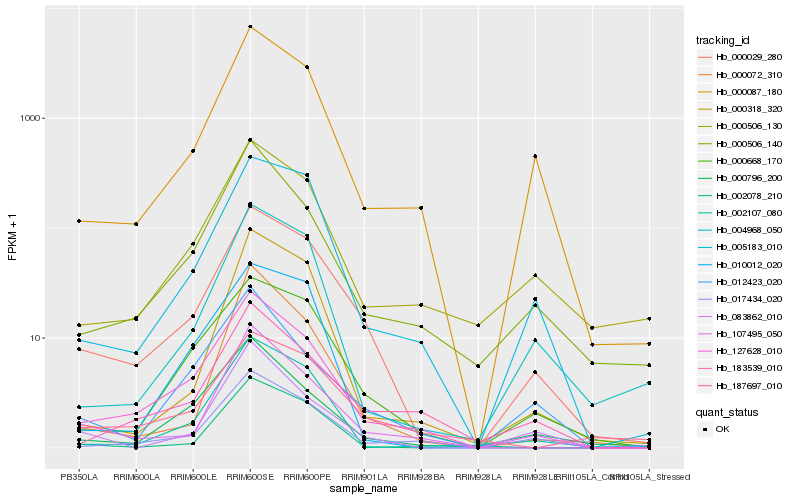
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 2 |
Hb_000668_170 |
0.1301864776 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 3 |
Hb_005183_010 |
0.1414431835 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 4 |
Hb_000087_180 |
0.148118523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_127628_010 |
0.1581374748 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_107495_050 |
0.1671497577 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 7 |
Hb_187697_010 |
0.167575524 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 8 |
Hb_017434_020 |
0.1761469468 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 9 |
Hb_004968_050 |
0.1806473946 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 10 |
Hb_000506_130 |
0.1822551095 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 11 |
Hb_002107_080 |
0.1843772327 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 12 |
Hb_000796_200 |
0.1870775446 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 13 |
Hb_000506_140 |
0.187198534 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 14 |
Hb_183539_010 |
0.189026557 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 15 |
Hb_000318_320 |
0.1916030313 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 16 |
Hb_083862_010 |
0.1954992885 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 17 |
Hb_012423_020 |
0.1998246178 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |
| 18 |
Hb_000072_310 |
0.1999389087 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 19 |
Hb_010012_020 |
0.2012633078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002078_210 |
0.2014856206 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |