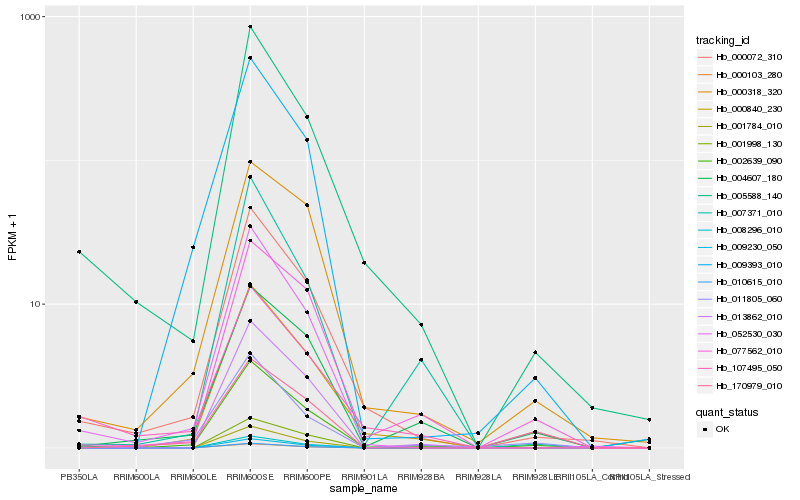
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 2 |
Hb_000840_230 |
0.0905492523 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 3 |
Hb_000072_310 |
0.0983375094 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 4 |
Hb_077562_010 |
0.1121962711 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 5 |
Hb_052530_030 |
0.1139063938 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 6 |
Hb_009393_010 |
0.1208350246 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 7 |
Hb_011805_060 |
0.1211932569 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_000318_320 |
0.1220912559 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 9 |
Hb_005588_140 |
0.123370866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_013862_010 |
0.1321291133 |
- |
- |
PREDICTED: uncharacterized protein LOC105650642 [Jatropha curcas] |
| 11 |
Hb_004607_180 |
0.1327546657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007371_010 |
0.1393819518 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 13 |
Hb_170979_010 |
0.1453312426 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 14 |
Hb_107495_050 |
0.1460477179 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 15 |
Hb_008296_010 |
0.1476207259 |
- |
- |
- |
| 16 |
Hb_009230_050 |
0.1476400879 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 17 |
Hb_001784_010 |
0.1476493252 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 18 |
Hb_010615_010 |
0.1478830092 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 19 |
Hb_000103_280 |
0.1482986527 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 20 |
Hb_001998_130 |
0.1486103647 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 35 family protein [Populus trichocarpa] |