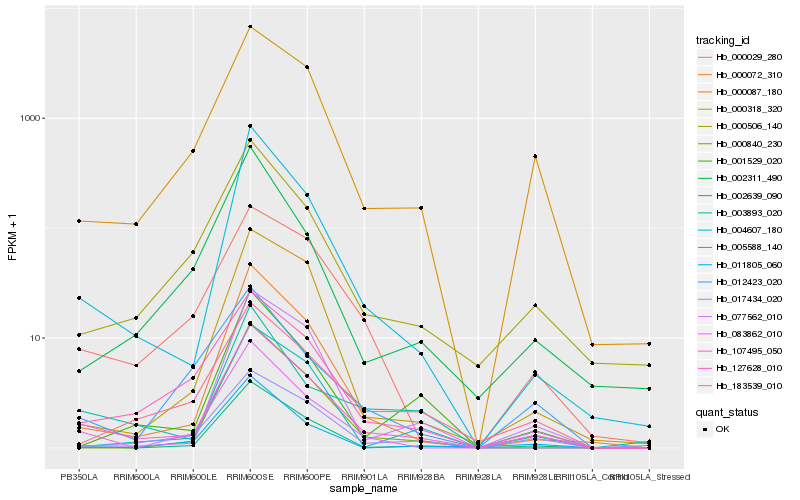
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107495_050 |
0.0 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 2 |
Hb_005588_140 |
0.1102131826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_083862_010 |
0.131175842 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 4 |
Hb_000072_310 |
0.1316526266 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 5 |
Hb_000087_180 |
0.1430176832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002639_090 |
0.1460477179 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 7 |
Hb_000506_140 |
0.1588565546 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 8 |
Hb_017434_020 |
0.1626516957 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 9 |
Hb_000840_230 |
0.16297527 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 10 |
Hb_000029_280 |
0.1671497577 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 11 |
Hb_183539_010 |
0.1732577539 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 12 |
Hb_000318_320 |
0.1768087807 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 13 |
Hb_127628_010 |
0.1770118314 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_001529_020 |
0.1772751546 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002311_490 |
0.183496947 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 16 |
Hb_003893_020 |
0.1842499639 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_004607_180 |
0.1850329517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011805_060 |
0.1850731521 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 19 |
Hb_077562_010 |
0.1863055877 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 20 |
Hb_012423_020 |
0.1889200494 |
- |
- |
hypothetical protein JCGZ_20730 [Jatropha curcas] |