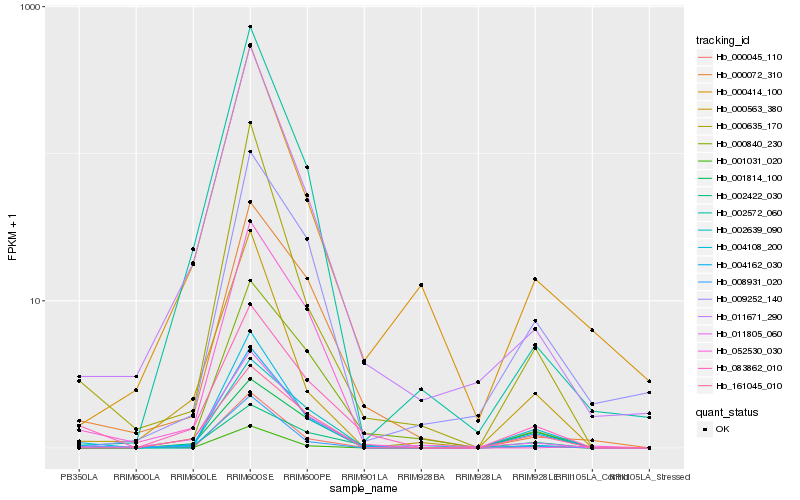
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_030 |
0.0 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Jatropha curcas] |
| 2 |
Hb_004108_200 |
0.1043955361 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_083862_010 |
0.1347907334 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 4 |
Hb_001031_020 |
0.1357488377 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 5 |
Hb_161045_010 |
0.1419663425 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 6 |
Hb_002422_030 |
0.1485043793 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 7 |
Hb_011805_060 |
0.1497535747 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_001814_100 |
0.1510973872 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 9 |
Hb_009252_140 |
0.1511522352 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 10 |
Hb_002639_090 |
0.153081444 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 11 |
Hb_000840_230 |
0.153450597 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 12 |
Hb_008931_020 |
0.1537531292 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 13 |
Hb_000045_110 |
0.1589858343 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 14 |
Hb_011671_290 |
0.1604319296 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_26299 [Jatropha curcas] |
| 15 |
Hb_000635_170 |
0.160873865 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000563_380 |
0.1719939017 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |
| 17 |
Hb_002572_060 |
0.1725930881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_052530_030 |
0.1774097129 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 19 |
Hb_000414_100 |
0.1808046069 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 20 |
Hb_000072_310 |
0.182133024 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |