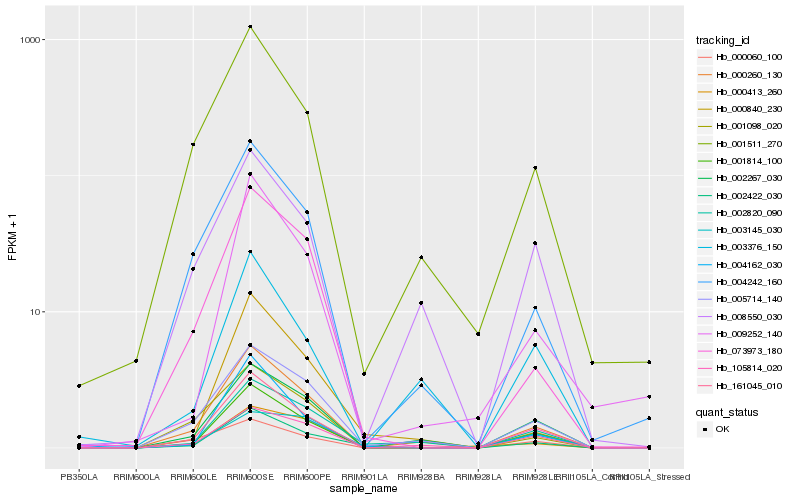
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001814_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105119066 [Populus euphratica] |
| 2 |
Hb_161045_010 |
0.0746902146 |
- |
- |
hypothetical protein JCGZ_15100 [Jatropha curcas] |
| 3 |
Hb_000260_130 |
0.118094501 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002422_030 |
0.1348066008 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 5 |
Hb_002267_030 |
0.1371189942 |
- |
- |
hypothetical protein JCGZ_24883 [Jatropha curcas] |
| 6 |
Hb_004162_030 |
0.1510973872 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Jatropha curcas] |
| 7 |
Hb_005714_140 |
0.1545076311 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_073973_180 |
0.1605751636 |
- |
- |
PREDICTED: WAT1-related protein At4g08290 isoform X1 [Populus euphratica] |
| 9 |
Hb_009252_140 |
0.1632777576 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 10 |
Hb_002820_090 |
0.1633449688 |
- |
- |
PREDICTED: WAT1-related protein At1g21890-like [Jatropha curcas] |
| 11 |
Hb_001511_270 |
0.1797497871 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 12 |
Hb_004242_160 |
0.1805806955 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000413_260 |
0.1847451228 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_001098_020 |
0.1871878868 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 15 |
Hb_008550_030 |
0.1881715895 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 16 |
Hb_105814_020 |
0.189317455 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 17 |
Hb_003376_150 |
0.192320616 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 18 |
Hb_003145_030 |
0.1939518019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000840_230 |
0.194333044 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 20 |
Hb_000060_100 |
0.1944181775 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |